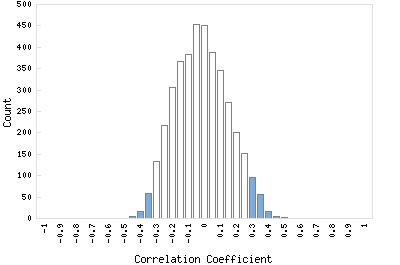
Correlation Catalog M. tuberculosis H37Rv: Rv1421 - conserved hypothetical protein
Distribution of gene correlation: Rv1421 vs. all genes
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0304c | PPE family protein |

|
 -0.41991 -0.41991
| |
| Rv3082c | araC/xylS-family virulence-regulating transcriptional regulator virS |

|
 -0.41893 -0.41893
| |
| Rv0150c | conserved hypothetical protein |

|
 -0.40473 -0.40473
| |
| Rv1552 | fumarate reductase flavoprotein subunit frdA |

|
 -0.4017 -0.4017
| |
| Rv2269c | hypothetical protein |

|
 -0.39606 -0.39606
| |
| Rv1291c | conserved secreted protein |

|
 -0.39485 -0.39485
| |
| Rv1888c | transmembrane protein |

|
 -0.38693 -0.38693
| |
| Rv2416c | enhanced intracellular survival protein eis |


|
 -0.3846 -0.3846
| |
| Rv0334 | alpha-D-glucose-1-phosphate thymidylyltransferase rmlA |

|
 -0.3834 -0.3834
| |
| Rv1926c | immunogenic protein mpt63 |

|
 -0.38141 -0.38141
| |
| Rv2716 | conserved hypothetical protein |

|
 -0.37645 -0.37645
| |
| Rv0344c | lipoprotein lpqJ |

|
 -0.36869 -0.36869
| |
| Rv1489c |

|
 -0.36726 -0.36726
| ||
| Rv0355c | PPE family protein |

|
 -0.36571 -0.36571
| |
| Rv1270c | lipoprotein lprA |

|
 -0.36556 -0.36556
| |
| Rv3108 | hypothetical protein |

|
 -0.35901 -0.35901
| |
| Rv1972 | MCE-associated membrane protein |

|
 -0.35836 -0.35836
| |
| Rv3110 | pterin-4-alpha-carbinolamine dehydratase moaB1 |

|
 -0.35587 -0.35587
| |
| Rv3128c | conserved hypothetical protein |

|
 -0.35462 -0.35462
| |
| Rv1876 | bacterioferritin bfrA |

|
 -0.35403 -0.35403
| |
| Rv0806c | UDP-glucose-4-epimerase cpsY |

|
 -0.35171 -0.35171
| |
| Rv2227 | conserved hypothetical protein |

|
 -0.34696 -0.34696
| |
| Rv0100 | conserved hypothetical protein |

|
 -0.34691 -0.34691
| |
| Rv0104 | conserved hypothetical protein |

|
 -0.346 -0.346
| |
| Rv3820c | polyketide synthase associated protein papA2 |

|
 -0.34419 -0.34419
| |
| Rv2872 | conserved hypothetical protein |

|
 -0.34346 -0.34346
| |
| Rv0008c | membrane protein |

|
 -0.34254 -0.34254
| |
| Rv0099 | fatty-acid-CoA ligase fadD10 |


|
 -0.33964 -0.33964
| |
| Rv0116c | conserved membrane protein |

|
 -0.3394 -0.3394
| |
| Rv1764 | transposase |

|
 -0.33798 -0.33798
| |
| Rv0224c | methyltransferase/methylase |


|
 -0.33555 -0.33555
| |
| Rv0130 | conserved hypothetical protein |

|
 -0.33482 -0.33482
| |
| Rv2332 | NAD-dependent malate oxidoreductase mez |

|
 -0.3348 -0.3348
| |
| Rv2253 | secreted protein |

|
 -0.3335 -0.3335
| |
| Rv1148c | conserved hypothetical protein |

|
 -0.33273 -0.33273
| |
| Rv1290c | conserved hypothetical protein |

|
 -0.33171 -0.33171
| |
| Rv1469 | cation transporter P-type ATPase D ctpD |

|
 -0.33037 -0.33037
| |
| Rv0573c | conserved hypothetical protein |

|
 -0.33016 -0.33016
| |
| Rv1490 | membrane protein |

|
 -0.32909 -0.32909
| |
| Rv0102 | conserved membrane protein |

|
 -0.32806 -0.32806
| |
| Rv3127 | conserved hypothetical protein |

|
 -0.32755 -0.32755
| |
| Rv0602c | two component system transcriptional regulator tcrA |


|
 -0.32395 -0.32395
| |
| Rv3637 | transposase |

|
 -0.32375 -0.32375
| |
| Rv1760 | conserved hypothetical protein |

|
 -0.32307 -0.32307
| |
| Rv0816c | thioredoxin thiX |


|
 -0.32261 -0.32261
| |
| Rv3359 | oxidoreductase |

|
 -0.32242 -0.32242
| |
| Rv2617c | transmembrane protein |

|
 -0.32181 -0.32181
| |
| Rv0575c | oxidoreductase |


|
 -0.32137 -0.32137
| |
| Rv2491 | conserved hypothetical protein |

|
 -0.32107 -0.32107
| |
| Rv3789 | conserved membrane protein |

|
 -0.32058 -0.32058
| |
| Rv1150 | transposase |

|
 -0.32012 -0.32012
| |
| Rv3111 | molybdenum cofactor biosynthesis protein C moaC1 |

|
 -0.31894 -0.31894
| |
| Rv3643 | hypothetical protein |

|
 -0.31893 -0.31893
| |
| Rv0096 | PPE family protein |

|
 -0.3189 -0.3189
| |
| Rv1499 | hypothetical protein |

|
 -0.31831 -0.31831
| |
| Rv3113 | phosphatase |

|
 -0.31794 -0.31794
| |
| Rv3174 | short-chain type dehydrogenase/reductase |


|
 -0.31746 -0.31746
| |
| Rv2546 | conserved hypothetical protein |

|
 -0.31686 -0.31686
| |
| Rv0117 | oxidative stress response regulatory protein oxyS |

|
 -0.31482 -0.31482
| |
| Rv3130c | conserved hypothetical protein |

|
 -0.31419 -0.31419
| |
| Rv3822 | conserved hypothetical protein |

|
 -0.31264 -0.31264
| |
| Rv1031 | potassium-transporting ATPase C chain kdpC |

|
 -0.31212 -0.31212
| |
| Rv1777 | cytochrome P450 144 cyp144 |

|
 -0.31187 -0.31187
| |
| Rv3714c | conserved hypothetical protein |

|
 -0.31051 -0.31051
| |
| Rv0873 | acyl-CoA dehydrogenase fadE10 |

|
 -0.31038 -0.31038
| |
| Rv3112 | molybdenum cofactor biosynthesis protein D moaD1 |

|
 -0.30986 -0.30986
| |
| Rv3018c | PPE family protein |

|
 -0.30974 -0.30974
| |
| Rv3379c | 1-deoxy-D-xylulose 5-phosphate synthase dxs2 |


|
 -0.30959 -0.30959
| |
| Rv0137c | peptide methionine sulfoxide reductase msrA |

|
 -0.30893 -0.30893
| |
| Rv0277c | conserved hypothetical protein |

|
 -0.30865 -0.30865
| |
| Rv1670 | conserved hypothetical protein |

|
 -0.30826 -0.30826
| |
| Rv1285 | sulfate adenylyltransferase subunit 2 cysD |


|
 -0.30566 -0.30566
| |
| Rv3126c | hypothetical protein |

|
 -0.3047 -0.3047
| |
| Rv0891c | transcriptional regulator |

|
 -0.30406 -0.30406
| |
| Rv0152c | PE family protein |

|
 -0.30343 -0.30343
| |
| Rv1918c | PPE family protein |

|
 -0.30311 -0.30311
| |
| Rv1978 | conserved hypothetical protein |


|
 -0.30299 -0.30299
| |
| Rv3845 | hypothetical protein |

|
 -0.30166 -0.30166
| |
| Rv1673c | conserved hypothetical protein |

|
 -0.30092 -0.30092
| |
| Rv1878 | glutamine synthetase glnA3 |

|
 -0.30011 -0.30011
|
Genes with positive correlation 
Categories Enriched  : E
: E
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0613c | hypothetical protein |

|
 0.53424 0.53424
| |
| Rv2613c | conserved hypothetical protein |



|
 0.51162 0.51162
| |
| Rv1603 | phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase hisA |

|
 0.48879 0.48879
| |
| Rv1596 | nicotinate-nucleotide pyrophosphatase nadC |

|
 0.48767 0.48767
| |
| Rv2611c | acyltransferase |

|
 0.46313 0.46313
| |
| Rv1595 | L-aspartate oxidase nadB |

|
 0.45195 0.45195
| |
| Rv1836c | conserved hypothetical protein |

|
 0.43851 0.43851
| |
| Rv2195 | Rieske iron-sulfur protein qcrA |

|
 0.435 0.435
| |
| Rv1449c | transketolase tkt |

|
 0.43233 0.43233
| |
| Rv3858c | NADH-dependent glutamate synthase small subunit gltD |


|
 0.42979 0.42979
| |
| Rv0018c | serine/threonine phosphatase ppp |

|
 0.42676 0.42676
| |
| Rv1173 | F420 biosynthesis protein fbiC |


|
 0.42443 0.42443
| |
| Rv1391 | DNA/pantothenate metabolism flavoprotein dfp |

|
 0.42207 0.42207
| |
| Rv2537c | 3-dehydroquinate dehydratase aroD |

|
 0.42009 0.42009
| |
| Rv0993 | UTP-glucose-1-phosphate uridylyltransferase galU |

|
 0.4189 0.4189
| |
| Rv2881c | membrane phosphatidate cytidylyltransferase cdsA |

|
 0.4148 0.4148
| |
| Rv3149 | NADH dehydrogenase I chain E nuoE |

|
 0.41033 0.41033
| |
| Rv1309 | ATP synthase gamma chain atpG |

|
 0.40982 0.40982
| |
| Rv0633c | hypothetical exported protein |

|
 0.40894 0.40894
| |
| Rv1559 | threonine dehydratase ilvA |

|
 0.40708 0.40708
| |
| Rv3626c | conserved hypothetical protein |

|
 0.40622 0.40622
| |
| Rv0247c | succinate dehydrogenase iron-sulfur subunit |

|
 0.40154 0.40154
| |
| Rv1597 | hypothetical protein |


|
 0.39993 0.39993
| |
| Rv0722 | 50S ribosomal protein L30 rpmD |

|
 0.39915 0.39915
| |
| Rv2109c | proteasome alpha subunit prcA |

|
 0.39491 0.39491
| |
| Rv1837c | malate synthase G glcB |

|
 0.39292 0.39292
| |
| Rv2477c | macrolide-transport ATP-binding protein ABC transporter |

|
 0.39087 0.39087
| |
| Rv3151 | NADH dehydrogenase I chain G nuoG |

|
 0.38937 0.38937
| |
| Rv1294 | homoserine dehydrogenase thrA |

|
 0.38934 0.38934
| |
| Rv3030 | conserved hypothetical protein |


|
 0.38767 0.38767
| |
| Rv1383 | carbamoyl-phosphate synthase small chain carA |


|
 0.38722 0.38722
| |
| Rv3607c | dihydroneopterin aldolase folB |

|
 0.38635 0.38635
| |
| Rv0710 | 30S ribosomal protein S17 rpsQ |

|
 0.38522 0.38522
| |
| Rv2936 | daunorubicin-dim-transport ATP-binding protein ABC transporter drrA |

|
 0.38513 0.38513
| |
| Rv1797 | conserved hypothetical protein |

|
 0.38448 0.38448
| |
| Rv3256c | conserved hypothetical protein |

|
 0.38412 0.38412
| |
| Rv2933 | phenolpthiocerol synthesis type-I polyketide synthase ppsC |

|
 0.38223 0.38223
| |
| Rv0663 | arylsulfatase atsD |

|
 0.38064 0.38064
| |
| Rv0931c | transmembrane serine/threonine-protein kinase D pknD |

|
 0.37861 0.37861
| |
| Rv2181 | conserved membrane protein |

|
 0.37744 0.37744
| |
| Rv1333 | hydrolase |


|
 0.37737 0.37737
| |
| Rv2883c | uridylate kinase pyrH |

|
 0.37688 0.37688
| |
| Rv1602 | amidotransferase hisH |

|
 0.3759 0.3759
| |
| Rv1310 | ATP synthase beta chain atpD |

|
 0.37482 0.37482
| |
| Rv2444c | ribonuclease E rne |

|
 0.37458 0.37458
| |
| Rv2367c | hypothetical protein |

|
 0.37431 0.37431
| |
| Rv3703c | conserved hypothetical protein |

|
 0.37379 0.37379
| |
| Rv3028c | electron transfer flavoprotein alpha subunit fixB |

|
 0.37138 0.37138
| |
| Rv3212 | conserved alanine and valine rich protein |

|
 0.37075 0.37075
| |
| Rv0721 | 30S ribosomal protein S5 rpsE |

|
 0.36898 0.36898
| |
| Rv0528 | conserved membrane protein |

|
 0.3675 0.3675
| |
| Rv3799c | propionyl-CoA carboxylase beta chain 4 accD4 |

|
 0.36737 0.36737
| |
| Rv1005c | para-aminobenzoate synthase component I pabD |


|
 0.36669 0.36669
| |
| Rv1340 | ribonuclease rphA |

|
 0.36622 0.36622
| |
| Rv2158c | UDP-N-acetylmuramoylalanyl-D-glutamate-2,6-diaminopimelat E ligase murE |

|
 0.36461 0.36461
| |
| Rv2859c | amidotransferase |

|
 0.36313 0.36313
| |
| Rv3341 | homoserine O-acetyltransferase metA |

|
 0.36284 0.36284
| |
| Rv2207 | nicotinate-nucleotide-dimethylbenzimidazol phosphoribosyltransferase cobT |

|
 0.36226 0.36226
| |
| Rv3800c | polyketide synthase pks13 |

|
 0.36206 0.36206
| |
| Rv0049 | conserved hypothetical protein |

|
 0.36154 0.36154
| |
| Rv0015c | transmembrane serine/threonine-protein kinase A pknA |

|
 0.36102 0.36102
| |
| Rv1007c | methionyl-tRNA synthetase metS |

|
 0.36047 0.36047
| |
| Rv2905 | alanine rich lipoprotein lppW |

|
 0.36036 0.36036
| |
| Rv1301 | conserved hypothetical protein |

|
 0.36026 0.36026
| |
| Rv2553c | conserved membrane protein |

|
 0.35936 0.35936
| |
| Rv1381 | dihydroorotase pyrC |

|
 0.35862 0.35862
| |
| Rv0513 | conserved membrane protein |

|
 0.35601 0.35601
| |
| Rv2196 | ubiquinol-cytochrome C reductase qcrB cytochrome B subunit |

|
 0.35555 0.35555
| |
| Rv3808c | bifunctional UDP-galactofuranosyl transferase glfT |

|
 0.35504 0.35504
| |
| Rv2923c | conserved hypothetical protein |

|
 0.35354 0.35354
| |
| Rv0512 | delta-aminolevulinic acid dehydratase hemB |

|
 0.35328 0.35328
| |
| Rv2703 | RNA polymerase sigma factor sigA |

|
 0.35257 0.35257
| |
| Rv2942 | transmembrane transport protein mmpL7 |

|
 0.35208 0.35208
| |
| Rv2163c | penicillin-binding membrane protein pbpB |

|
 0.35196 0.35196
| |
| Rv2157c | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanyl ligase murF |

|
 0.35183 0.35183
| |
| Rv1328 | glycogen phosphorylase glgP |

|
 0.35148 0.35148
| |
| Rv0509 | glutamyl-tRNA reductase hemA |

|
 0.35135 0.35135
| |
| Rv1324 | thioredoxin |

|
 0.3494 0.3494
| |
| Rv0207c | conserved hypothetical protein |

|
 0.34921 0.34921
| |
| Rv3805c | conserved membrane protein |

|
 0.34762 0.34762
| |
| Rv1635c | conserved membrane protein |

|
 0.34756 0.34756
| |
| Rv1411c | lipoprotein lprG |

|
 0.34727 0.34727
| |
| Rv1615 | hypothetical membrane protein |

|
 0.34568 0.34568
| |
| Rv3709c | aspartokinase ask |

|
 0.34534 0.34534
| |
| Rv1601 | imidazole glycerol-phosphate dehydratase hisB |

|
 0.34477 0.34477
| |
| Rv3285 | bifunctional acetyl-/propionyl-coenzyme A carboxylase alpha chain accA3 |

|
 0.34458 0.34458
| |
| Rv0632c | enoyl-CoA hydratase echA3 |

|
 0.34445 0.34445
| |
| Rv1223 | serine protease htrA |

|
 0.34408 0.34408
| |
| Rv2868c | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase gcpE |

|
 0.34371 0.34371
| |
| Rv3284 | conserved hypothetical protein |

|
 0.34347 0.34347
| |
| Rv1693 | conserved hypothetical protein |

|
 0.34153 0.34153
| |
| Rv1605 | cyclase hisF |

|
 0.34147 0.34147
| |
| Rv1315 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase murA |

|
 0.3414 0.3414
| |
| Rv2970c | lipase/esterase lipN |

|
 0.34109 0.34109
| |
| Rv0803 | phosphoribosylformylglycinamidine synthase II purL |

|
 0.34066 0.34066
| |
| Rv3257c | phosphomannomutase pmmA |

|
 0.33937 0.33937
| |
| Rv2932 | phenolpthiocerol synthesis type-I polyketide synthase ppsB |

|
 0.33876 0.33876
| |
| Rv2753c | dihydrodipicolinate synthase dapA |


|
 0.33677 0.33677
| |
| Rv1311 | ATP synthase epsilon chain atpC |

|
 0.33653 0.33653
| |
| Rv2164c | conserved proline rich membrane protein |

|
 0.33627 0.33627
| |
| Rv2245 | 3-oxoacyl-[acyl-carrier protein] synthase 1 kasA |


|
 0.33548 0.33548
| |
| Rv1600 | histidinol-phosphate aminotransferase hisC1 |

|
 0.33456 0.33456
| |
| Rv1483 | 3-oxoacyl-[acyl-carrier protein] reductase fabG1 |


|
 0.33372 0.33372
| |
| Rv0050 | bifunctional penicillin-binding protein ponA1 |

|
 0.33321 0.33321
| |
| Rv1677 | lipoprotein dsbF |


|
 0.33264 0.33264
| |
| Rv1165 | GTP-binding translation elongation factor typA |

|
 0.33075 0.33075
| |
| Rv1025 | conserved hypothetical protein |

|
 0.33015 0.33015
| |
| Rv0525 | conserved hypothetical protein |

|
 0.32953 0.32953
| |
| Rv0289 | conserved hypothetical protein |

|
 0.32837 0.32837
| |
| Rv1415 | riboflavin biosynthesis protein ribA2 |

|
 0.32804 0.32804
| |
| Rv3157 | NADH dehydrogenase I chain M nuoM |

|
 0.32754 0.32754
| |
| Rv2922c | chromosome partition protein smc |

|
 0.3271 0.3271
| |
| Rv0587 | hypothetical membrane protein yrbE2A |

|
 0.32688 0.32688
| |
| Rv1437 | phosphoglycerate kinase pgk |

|
 0.32664 0.32664
| |
| Rv1565c | conserved membrane protein |

|
 0.32552 0.32552
| |
| Rv0709 | 50S ribosomal protein L29 rpmC |

|
 0.32356 0.32356
| |
| Rv0365c | conserved hypothetical protein |

|
 0.32308 0.32308
| |
| Rv0005 | DNA gyrase subunit B gyrB |

|
 0.3228 0.3228
| |
| Rv1484 | NADH-dependent enoyl-[acyl-carrier-protein] reductase inhA |

|
 0.32186 0.32186
| |
| Rv3813c | conserved hypothetical protein |

|
 0.32138 0.32138
| |
| Rv1785c | cytochrome P450 143 cyp143 |

|
 0.32117 0.32117
| |
| Rv1011 | 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase ispE |

|
 0.32059 0.32059
| |
| Rv3039c | enoyl-CoA hydratase echA17 |

|
 0.32042 0.32042
| |
| Rv1382 | hypothetical exported protein |

|
 0.31922 0.31922
| |
| Rv1422 | conserved hypothetical protein |

|
 0.31838 0.31838
| |
| Rv1696 | DNA repair protein recN |

|
 0.31778 0.31778
| |
| Rv1655 | acetylornithine aminotransferase argD |

|
 0.31717 0.31717
| |
| Rv0410c | serine/threonine-protein kinase pknG |

|
 0.31673 0.31673
| |
| Rv3024c | tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase trmU |

|
 0.31594 0.31594
| |
| Rv2931 | phenolpthiocerol synthesis type-I polyketide synthase ppsA |

|
 0.31593 0.31593
| |
| Rv1859 | molybdenum-transport ATP-binding protein ABC transporter modC |

|
 0.31558 0.31558
| |
| Rv1495 | conserved hypothetical protein |

|
 0.31523 0.31523
| |
| Rv2152c | UDP-N-acetylmuramate-alanine ligase murC |

|
 0.31458 0.31458
| |
| Rv1097c | glycine and proline rich membrane protein |

|
 0.31391 0.31391
| |
| Rv0019c | conserved hypothetical protein |

|
 0.31359 0.31359
| |
| Rv2365c | conserved hypothetical protein |

|
 0.3131 0.3131
| |
| Rv0861c | DNA helicase ercc3 |

|
 0.31234 0.31234
| |
| Rv0932c | periplasmic phosphate-binding lipoprotein pstS2 |

|
 0.31227 0.31227
| |
| Rv3877 | conserved membrane protein |

|
 0.31223 0.31223
| |
| Rv1701 | integrase |

|
 0.31167 0.31167
| |
| Rv3283 | thiosulfate sulfurtransferase sseA |

|
 0.31101 0.31101
| |
| Rv1233c | conserved membrane protein |

|
 0.31095 0.31095
| |
| Rv0183 | lysophospholipase |

|
 0.31067 0.31067
| |
| Rv0299 | hypothetical protein |

|
 0.30977 0.30977
| |
| Rv0733 | adenylate kinase adk |

|
 0.30906 0.30906
| |
| Rv3003c | acetolactate synthase large subunit ilvB1 |
![COG0028: (EH) Thiamine pyrophosphate-requiring enzymes [acetolactate synthase; pyruvate dehydrogenase (cytochrome); glyoxylate carboligase; phosphonopyruvate decarboxylase]](supportFiles/E.png)
![COG0028: (EH) Thiamine pyrophosphate-requiring enzymes [acetolactate synthase; pyruvate dehydrogenase (cytochrome); glyoxylate carboligase; phosphonopyruvate decarboxylase]](supportFiles/H.png)
|
 0.30827 0.30827
| |
| Rv3303c | dihydrolipoamide dehydrogenase lpdA |

|
 0.30688 0.30688
| |
| Rv0421c | conserved hypothetical protein |

|
 0.30685 0.30685
| |
| Rv3605c | conserved secreted protein |

|
 0.30595 0.30595
| |
| Rv0283 | conserved membrane protein |

|
 0.30563 0.30563
| |
| Rv0411c | glutamine-binding lipoprotein glnH |

|
 0.30554 0.30554
| |
| Rv2447c | folylpolyglutamate synthase protein folC |

|
 0.30548 0.30548
| |
| Rv1480 | conserved hypothetical protein |

|
 0.30543 0.30543
| |
| Rv2587c | protein-export membrane protein secD |

|
 0.30532 0.30532
| |
| Rv0809 | phosphoribosylformylglycinamidine cyclo-ligase purM |

|
 0.3048 0.3048
| |
| Rv1539 | lipoprotein signal peptidase lspA |

|
 0.30467 0.30467
| |
| Rv1121 | glucose-6-phosphate 1-dehydrogenase zwf1 |

|
 0.30429 0.30429
| |
| Rv1229c | hypothetical protein mrp |

|
 0.30378 0.30378
| |
| Rv1003 | conserved hypothetical protein |

|
 0.30371 0.30371
| |
| Rv0423c | thiamine biosynthesis protein thiC |

|
 0.3034 0.3034
| |
| Rv2151c | cell division protein ftsQ |

|
 0.3033 0.3033
| |
| Rv2783c | bifunctional polyribonucleotide nucleotidyltransferase gpsI |

|
 0.30324 0.30324
| |
| Rv1417 | conserved membrane protein |

|
 0.30324 0.30324
| |
| Rv0273c | transcriptional regulator |

|
 0.30289 0.30289
| |
| Rv2773c | dihydrodipicolinate reductase dapB |

|
 0.30287 0.30287
| |
| Rv2538c | 3-dehydroquinate synthase aroB |

|
 0.30268 0.30268
| |
| Rv2064 | cobalamin biosynthesis protein cobG |

|
 0.30258 0.30258
| |
| Rv3336c | tryptophanyl-tRNA synthetase trpS |

|
 0.30225 0.30225
| |
| Rv0511 | uroporphyrin-III C-methyltransferase hemD |

|
 0.3019 0.3019
| |
| Rv2215 | pyruvate dehydrogenase E2 component sucB |

|
 0.30189 0.30189
| |
| Rv3869 | conserved membrane protein |

|
 0.30079 0.30079
| |
| Rv3868 | conserved hypothetical protein |

|
 0.30009 0.30009
| |
| Rv1659 | argininosuccinate lyase argH |

|
 0.30008 0.30008
|