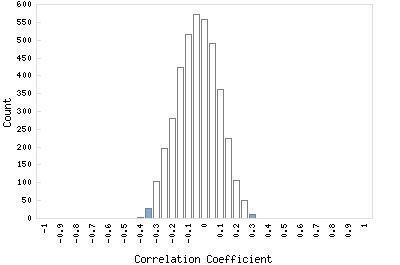
Correlation Catalog M. tuberculosis H37Rv: Rv2776c - oxidoreductase
Distribution of gene correlation: Rv2776c vs. all genes
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0704 | 50S ribosomal protein L2 rplB |

|
 -0.3541 -0.3541
| |
| Rv0705 | 30S ribosomal protein S19 rpsS |

|
 -0.35248 -0.35248
| |
| Rv0237 | lipoprotein lpqI |

|
 -0.34686 -0.34686
| |
| Rv0555 | bifunctional menaquinone biosynthesis protein menD |

|
 -0.34063 -0.34063
| |
| Rv0545c | low affinity inorganic phosphate transporter membrane protein pitA |

|
 -0.34046 -0.34046
| |
| Rv0429c | polypeptide deformylase def |

|
 -0.33772 -0.33772
| |
| Rv0567 | methyltransferase/methylase |


|
 -0.33524 -0.33524
| |
| Rv0249c | succinate dehydrogenase membrane anchor subunit |

|
 -0.33481 -0.33481
| |
| Rv0546c | conserved hypothetical protein |

|
 -0.32796 -0.32796
| |
| Rv2113 | membrane protein |

|
 -0.3278 -0.3278
| |
| Rv1201c | transferase |

|
 -0.32634 -0.32634
| |
| Rv0537c | membrane protein |

|
 -0.32556 -0.32556
| |
| Rv0023 | transcriptional regulator |

|
 -0.3234 -0.3234
| |
| Rv0718 | 30S ribosomal protein S8 rpsH |

|
 -0.32114 -0.32114
| |
| Rv1379 | pyrimidine operon regulatory protein pyrR |

|
 -0.31767 -0.31767
| |
| Rv0482 | UDP-N-acetylenolpyruvoylglucosamine reductase murB |

|
 -0.31672 -0.31672
| |
| Rv0707 | 30S ribosomal protein S3 rpsC |

|
 -0.31448 -0.31448
| |
| Rv0703 | 50S ribosomal protein L23 rplW |

|
 -0.31347 -0.31347
| |
| Rv0720 | 50S ribosomal protein L18 rplR |

|
 -0.31234 -0.31234
| |
| Rv2523c | holo-[acyl-carrier protein] synthase acpS |

|
 -0.31037 -0.31037
| |
| Rv0643c | methoxy mycolic acid synthase 3 mmaA3 |

|
 -0.30937 -0.30937
| |
| Rv0701 | 50S ribosomal protein L3 rplC |

|
 -0.30932 -0.30932
| |
| Rv0155 | NAD(P) transhydrogenase alpha subunit pntAa |

|
 -0.30698 -0.30698
| |
| Rv0030 | hypothetical protein |

|
 -0.30601 -0.30601
| |
| Rv0438c | molybdopterin biosynthesis protein moeA2 |

|
 -0.30383 -0.30383
| |
| Rv0708 | 50S ribosomal protein L16 rplP |

|
 -0.30368 -0.30368
| |
| Rv3737 | conserved membrane protein |

|
 -0.30363 -0.30363
| |
| Rv0719 | 50S ribosomal protein L6 rplF |

|
 -0.30269 -0.30269
| |
| Rv2068c | class A beta-lactamase blaC |

|
 -0.30117 -0.30117
| |
| Rv1591 | transmembrane protein |

|
 -0.30076 -0.30076
|
Genes with positive correlation 
Categories Enriched  : None
: None
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv1958c | hypothetical protein |

|
 0.40875 0.40875
| |
| Rv2359 | ferric uptake regulation protein furB |

|
 0.35461 0.35461
| |
| Rv0786c | conserved hypothetical protein |

|
 0.3482 0.3482
| |
| Rv3238c | conserved membrane protein |

|
 0.3475 0.3475
| |
| Rv2469c | conserved hypothetical protein |

|
 0.33423 0.33423
| |
| Rv2342 | conserved hypothetical protein |

|
 0.32676 0.32676
| |
| Rv2575 | conserved glycine rich membrane protein |

|
 0.32623 0.32623
| |
| Rv0937c | conserved hypothetical protein |

|
 0.31845 0.31845
| |
| Rv1736c | nitrate reductase narX |

|
 0.31322 0.31322
| |
| Rv3502c | short-chain type dehydrogenase/reductase |


|
 0.31288 0.31288
| |
| Rv1829 | conserved hypothetical protein |

|
 0.31249 0.31249
| |
| Rv2495c | dihydrolipoamide S-acetyltransferase E2 component pdhC |

|
 0.30367 0.30367
|