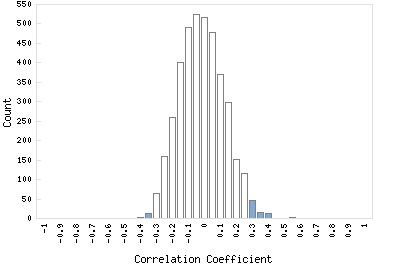
Correlation Catalog M. tuberculosis H37Rv: Rv1652 - N-acetl-gamma-glutamyl-phoshate reductase argC
Distribution of gene correlation: Rv1652 vs. all genes
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv1402 | primosomal protein N priA |

|
 -0.3873 -0.3873
| |
| Rv1271c | conserved secreted protein |

|
 -0.36052 -0.36052
| |
| Rv2716 | conserved hypothetical protein |

|
 -0.34756 -0.34756
| |
| Rv2817c | conserved hypothetical protein |

|
 -0.34754 -0.34754
| |
| Rv1873 | conserved hypothetical protein |

|
 -0.34408 -0.34408
| |
| Rv3753c | conserved hypothetical protein |

|
 -0.32302 -0.32302
| |
| Rv2633c | hypothetical protein |

|
 -0.32124 -0.32124
| |
| Rv0403c | membrane protein mmpS1 |

|
 -0.31997 -0.31997
| |
| Rv3849 | conserved hypothetical protein |

|
 -0.31576 -0.31576
| |
| Rv3108 | hypothetical protein |

|
 -0.31486 -0.31486
| |
| Rv1549 | fatty-acid-CoA ligase fadD11_1 |

|
 -0.31009 -0.31009
| |
| Rv2529 | hypothetical protein |

|
 -0.30528 -0.30528
| |
| Rv3847 | hypothetical protein |

|
 -0.30458 -0.30458
| |
| Rv1707 | conserved membrane protein |

|
 -0.30445 -0.30445
| |
| Rv0935 | phosphate-transport membrane ABC transporter pstC1 |

|
 -0.30416 -0.30416
| |
| Rv3097c | PE-PGRS family protein |

|
 -0.30053 -0.30053
|
Genes with positive correlation 
Categories Enriched  : None
: None
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv1654 | acetylglutamate kinase argB |

|
 0.67313 0.67313
| |
| Rv1656 | ornithine carbamoyltransferase argF |

|
 0.57617 0.57617
| |
| Rv1653 | glutamate n-acetyltransferase argJ |

|
 0.55705 0.55705
| |
| Rv3306c | amidohydrolase amiB1 |

|
 0.44705 0.44705
| |
| Rv0967 | conserved hypothetical protein |

|
 0.44434 0.44434
| |
| Rv0688 | ferredoxin reductase |

|
 0.43184 0.43184
| |
| Rv0982 | two component system sensor kinase mprB |

|
 0.42691 0.42691
| |
| Rv0975c | acyl-CoA dehydrogenase fadE13 |

|
 0.42264 0.42264
| |
| Rv2992c | glutamyl-tRNA synthetase gltS |

|
 0.42063 0.42063
| |
| Rv1657 | arginine repressor argR |

|
 0.41857 0.41857
| |
| Rv2078 | hypothetical protein |

|
 0.41378 0.41378
| |
| Rv1382 | hypothetical exported protein |

|
 0.41054 0.41054
| |
| Rv0484c | short-chain type oxidoreductase |

|
 0.40805 0.40805
| |
| Rv1292 | arginyl-tRNA synthetase argS |

|
 0.40628 0.40628
| |
| Rv1638 | excinuclease ABC subunit A uvrA |

|
 0.40272 0.40272
| |
| Rv0991c | conserved serine rich protein |

|
 0.39747 0.39747
| |
| Rv0356c | conserved hypothetical protein |

|
 0.39541 0.39541
| |
| Rv2614c | threonyl-tRNA synthetase thrS |

|
 0.38089 0.38089
| |
| Rv2448c | valyl-tRNA synthase protein valS |

|
 0.38052 0.38052
| |
| Rv1480 | conserved hypothetical protein |

|
 0.37857 0.37857
| |
| Rv0486 | mannosyltransferase |

|
 0.3767 0.3767
| |
| Rv1465 | nitrogen fixation related protein |

|
 0.37533 0.37533
| |
| Rv0512 | delta-aminolevulinic acid dehydratase hemB |

|
 0.37382 0.37382
| |
| Rv3014c | NAD-dependent DNA ligase ligA |

|
 0.37121 0.37121
| |
| Rv1406 | methionyl-tRNA formyltransferase fmt |

|
 0.36373 0.36373
| |
| Rv1294 | homoserine dehydrogenase thrA |

|
 0.36345 0.36345
| |
| Rv2678c | uroporphyrinogen decarboxylase hemE |

|
 0.35842 0.35842
| |
| Rv2112c | conserved hypothetical protein |

|
 0.35657 0.35657
| |
| Rv1462 | conserved hypothetical protein |

|
 0.35291 0.35291
| |
| Rv3710 | 2-isopropylmalate synthase leuA |

|
 0.35125 0.35125
| |
| Rv0766c | cytochrome P450 123 cyp123 |

|
 0.34676 0.34676
| |
| Rv3030 | conserved hypothetical protein |


|
 0.34595 0.34595
| |
| Rv0979c | hypothetical protein |

|
 0.34473 0.34473
| |
| Rv1854c | NADH dehydrogenase ndh |

|
 0.34457 0.34457
| |
| Rv0752c | acyl-CoA dehydrogenase fadE9 |

|
 0.3443 0.3443
| |
| Rv2064 | cobalamin biosynthesis protein cobG |

|
 0.34242 0.34242
| |
| Rv0568 | cytochrome P450 135B1 cyp135B1 |

|
 0.34131 0.34131
| |
| Rv0353 | merR-family heat shock protein transcriptional repressor hspR |

|
 0.34087 0.34087
| |
| Rv0410c | serine/threonine-protein kinase pknG |

|
 0.34056 0.34056
| |
| Rv0978c | PE-PGRS family protein |

|
 0.33949 0.33949
| |
| Rv1463 | ABC transporter ATP-binding protein |

|
 0.33939 0.33939
| |
| Rv0326 | hypothetical protein |


|
 0.33711 0.33711
| |
| Rv2620c | conserved membrane protein |

|
 0.33614 0.33614
| |
| Rv3800c | polyketide synthase pks13 |

|
 0.33513 0.33513
| |
| Rv1466 | conserved hypothetical protein |

|
 0.33374 0.33374
| |
| Rv1464 | cysteine desulfurase csd |

|
 0.33069 0.33069
| |
| Rv1476 | membrane protein |

|
 0.33023 0.33023
| |
| Rv1342c | conserved membrane protein |

|
 0.32695 0.32695
| |
| Rv0351 | chaperone grpE |

|
 0.32528 0.32528
| |
| Rv0350 | chaperone protein dnaK |

|
 0.32366 0.32366
| |
| Rv2470 | oxygen-binding protein globin glbO |

|
 0.32263 0.32263
| |
| Rv3024c | tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase trmU |

|
 0.3219 0.3219
| |
| Rv1832 | glycine dehydrogenase gcvB |

|
 0.31986 0.31986
| |
| Rv0294 | trans-aconitate methyltransferase tam |


|
 0.31844 0.31844
| |
| Rv3801c | fatty-acid-CoA ligase fadD32 |


|
 0.31826 0.31826
| |
| Rv3400 | hydrolase |

|
 0.31816 0.31816
| |
| Rv1102c | conserved hypothetical protein |

|
 0.31633 0.31633
| |
| Rv0818 | transcriptional regulator |


|
 0.31577 0.31577
| |
| Rv1602 | amidotransferase hisH |

|
 0.31477 0.31477
| |
| Rv1086 | short-chain Z-isoprenyl diphosphate synthase |

|
 0.31369 0.31369
| |
| Rv3536c | hydratase |

|
 0.31324 0.31324
| |
| Rv1296 | homoserine kinase thrB |

|
 0.31279 0.31279
| |
| Rv3626c | conserved hypothetical protein |

|
 0.31276 0.31276
| |
| Rv0987 | adhesion component transport transmembrane protein ABC transporter |

|
 0.31177 0.31177
| |
| Rv1982c | conserved hypothetical protein |

|
 0.30869 0.30869
| |
| Rv1120c | conserved hypothetical protein |

|
 0.30781 0.30781
| |
| Rv2852c | malate:quinone oxidoreductase mqo |

|
 0.30643 0.30643
| |
| Rv2110c | proteasome beta subunit prcB |

|
 0.30581 0.30581
| |
| Rv1260 | oxidoreductase |


|
 0.30562 0.30562
| |
| Rv0245 | oxidoreductase |

|
 0.30486 0.30486
| |
| Rv3256c | conserved hypothetical protein |

|
 0.30475 0.30475
| |
| Rv2454c | oxidoreductase beta subunit |

|
 0.30336 0.30336
| |
| Rv3484 | hypothetical protein cpsA |

|
 0.30335 0.30335
| |
| Rv0538 | conserved membrane protein |

|
 0.30317 0.30317
| |
| Rv0416 | hypothetical protein thiS |

|
 0.30141 0.30141
| |
| Rv1072 | conserved membrane protein |

|
 0.30105 0.30105
| |
| Rv2252 | conserved hypothetical protein |

|
 0.30072 0.30072
|