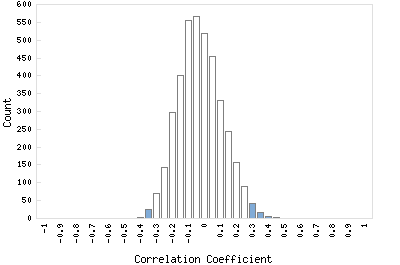
Correlation Catalog M. tuberculosis H37Rv: Rv0504c - conserved hypothetical protein
Distribution of gene correlation: Rv0504c vs. all genes
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv2160c | conserved hypothetical protein |

|
 -0.40156 -0.40156
| |
| Rv3625c | cell cycle protein mesJ |

|
 -0.38037 -0.38037
| |
| Rv3201c | ATP-dependent DNA helicase |

|
 -0.36864 -0.36864
| |
| Rv3540c | lipid transfer protein or keto acyl-CoA thiolase ltp2 |

|
 -0.34581 -0.34581
| |
| Rv3605c | conserved secreted protein |

|
 -0.343 -0.343
| |
| Rv2879c | conserved hypothetical protein |

|
 -0.33577 -0.33577
| |
| Rv2793c | tRNA pseudouridine synthase B truB |

|
 -0.33417 -0.33417
| |
| Rv1853 | urease accessory protein ureD |

|
 -0.33186 -0.33186
| |
| Rv1417 | conserved membrane protein |

|
 -0.33144 -0.33144
| |
| Rv3579c | tRNA/rRNA methyltransferase |

|
 -0.32904 -0.32904
| |
| Rv2849c | cob(I)alamin adenosyltransferase cobO |

|
 -0.3218 -0.3218
| |
| Rv2851c | conserved hypothetical protein |

|
 -0.31668 -0.31668
| |
| Rv3821 | conserved membrane protein |

|
 -0.31239 -0.31239
| |
| Rv2726c | diaminopimelate epimerase dapF |

|
 -0.31193 -0.31193
| |
| Rv3508 | PE-PGRS family protein |

|
 -0.30933 -0.30933
| |
| Rv2539c | shikimate kinase aroK |

|
 -0.30858 -0.30858
| |
| Rv2788 | transcriptional repressor sirR |

|
 -0.30797 -0.30797
| |
| Rv2164c | conserved proline rich membrane protein |

|
 -0.30685 -0.30685
| |
| Rv3243c | hypothetical protein |

|
 -0.30611 -0.30611
| |
| Rv2010 | conserved hypothetical protein |

|
 -0.30609 -0.30609
| |
| Rv2552c | shikimate 5-dehydrogenase aroE |

|
 -0.30577 -0.30577
| |
| Rv1713 | GTP-binding protein engA |

|
 -0.30565 -0.30565
| |
| Rv1797 | conserved hypothetical protein |

|
 -0.30456 -0.30456
| |
| Rv2417c | conserved hypothetical protein |

|
 -0.30319 -0.30319
| |
| Rv2528c | restriction system protein mrr |

|
 -0.30152 -0.30152
| |
| Rv1693 | conserved hypothetical protein |

|
 -0.30149 -0.30149
| |
| Rv2811 | conserved hypothetical protein |

|
 -0.30121 -0.30121
|
Genes with positive correlation 
Categories Enriched  : None
: None
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0338c | iron-sulfur-binding reductase |

|
 0.56379 0.56379
| |
| Rv0344c | lipoprotein lpqJ |

|
 0.4716 0.4716
| |
| Rv0306 | oxidoreductase |

|
 0.45298 0.45298
| |
| Rv0334 | alpha-D-glucose-1-phosphate thymidylyltransferase rmlA |

|
 0.44223 0.44223
| |
| Rv1290c | conserved hypothetical protein |

|
 0.44198 0.44198
| |
| Rv0518 | hypothetical exported protein |

|
 0.43828 0.43828
| |
| Rv0770 | dehydrogenase/reductase |

|
 0.43217 0.43217
| |
| Rv0566c | conserved hypothetical protein |

|
 0.4278 0.4278
| |
| Rv0296c | sulfatase |

|
 0.41731 0.41731
| |
| Rv0502 | conserved hypothetical protein |

|
 0.40552 0.40552
| |
| Rv1722 | carboxylase |

|
 0.39979 0.39979
| |
| Rv2958c | glycosyltransferase |


|
 0.3961 0.3961
| |
| Rv1674c | transcriptional regulator |

|
 0.39514 0.39514
| |
| Rv0156 | NAD(P) transhydrogenase alpha subunit pntAb |

|
 0.39367 0.39367
| |
| Rv0891c | transcriptional regulator |

|
 0.38939 0.38939
| |
| Rv1511 | GDP-D-mannose dehydratase gmdA |

|
 0.38702 0.38702
| |
| Rv3820c | polyketide synthase associated protein papA2 |

|
 0.38237 0.38237
| |
| Rv0130 | conserved hypothetical protein |

|
 0.38173 0.38173
| |
| Rv3446c | hypothetical alanine and valine rich protein |

|
 0.37684 0.37684
| |
| Rv3012c | glutamyl-tRNA(gln) amidotransferase subunit C gatC |

|
 0.36531 0.36531
| |
| Rv0150c | conserved hypothetical protein |

|
 0.3637 0.3637
| |
| Rv2290 | lipoprotein lppO |

|
 0.36261 0.36261
| |
| Rv0500 | pyrroline-5-carboxylate reductase proC |

|
 0.35925 0.35925
| |
| Rv2275 | conserved hypothetical protein |

|
 0.35306 0.35306
| |
| Rv0358 | conserved hypothetical protein |

|
 0.35147 0.35147
| |
| Rv2606c | pyridoxine biosynthesis protein |

|
 0.3512 0.3512
| |
| Rv0546c | conserved hypothetical protein |

|
 0.34583 0.34583
| |
| Rv1752 | conserved hypothetical protein |

|
 0.34486 0.34486
| |
| Rv0956 | 5-phosphoribosylglycinamide formyltransferase purN |

|
 0.34224 0.34224
| |
| Rv0316 | muconolactone isomerase |

|
 0.33933 0.33933
| |
| Rv2273 | conserved membrane protein |

|
 0.33728 0.33728
| |
| Rv0356c | conserved hypothetical protein |

|
 0.33718 0.33718
| |
| Rv0494 | transcriptional regulator, gntR-family |

|
 0.33589 0.33589
| |
| Rv2872 | conserved hypothetical protein |

|
 0.33543 0.33543
| |
| Rv1092c | pantothenate kinase coaA |

|
 0.33466 0.33466
| |
| Rv2804c | hypothetical protein |

|
 0.33375 0.33375
| |
| Rv0164 | conserved hypothetical protein |

|
 0.33277 0.33277
| |
| Rv1031 | potassium-transporting ATPase C chain kdpC |

|
 0.33248 0.33248
| |
| Rv0322 | UDP-glucose 6-dehydrogenase udgA |

|
 0.33083 0.33083
| |
| Rv0394c | secreted protein |

|
 0.32974 0.32974
| |
| Rv1876 | bacterioferritin bfrA |

|
 0.32785 0.32785
| |
| Rv1456c | antibiotic-transport membrane ABC transporter |

|
 0.32669 0.32669
| |
| Rv0330c | hypothetical protein |

|
 0.32614 0.32614
| |
| Rv1924c | hypothetical protein |

|
 0.32324 0.32324
| |
| Rv0740 | conserved hypothetical protein |

|
 0.32103 0.32103
| |
| Rv0948c | conserved hypothetical protein |

|
 0.31792 0.31792
| |
| Rv2856 | nickel-transport membrane protein nicT |

|
 0.3171 0.3171
| |
| Rv3634c | UDP-glucose 4-epimerase galE1 |


|
 0.31704 0.31704
| |
| Rv2416c | enhanced intracellular survival protein eis |


|
 0.31615 0.31615
| |
| Rv0354c | PPE family protein |

|
 0.31597 0.31597
| |
| Rv0277c | conserved hypothetical protein |

|
 0.31571 0.31571
| |
| Rv1270c | lipoprotein lprA |

|
 0.31497 0.31497
| |
| Rv3669 | conserved membrane protein |

|
 0.31386 0.31386
| |
| Rv2018 | conserved hypothetical protein |

|
 0.31268 0.31268
| |
| Rv0759c | conserved hypothetical protein |



|
 0.31258 0.31258
| |
| Rv1390 | DNA-directed RNA polymerase omega chain rpoZ |

|
 0.30954 0.30954
| |
| Rv0304c | PPE family protein |

|
 0.30933 0.30933
| |
| Rv3789 | conserved membrane protein |

|
 0.30763 0.30763
| |
| Rv0160c | PE family protein |

|
 0.30694 0.30694
| |
| Rv0380c | RNA methyltransferase |

|
 0.30642 0.30642
| |
| Rv1388 | integration host factor mihF |

|
 0.30588 0.30588
| |
| Rv0256c | PPE family protein |

|
 0.30422 0.30422
| |
| Rv3392c | cyclopropane-fatty-acyl-phospholipid synthase 1 cmaA1 |

|
 0.30415 0.30415
| |
| Rv0372c | conserved hypothetical protein |

|
 0.30234 0.30234
| |
| Rv0983 | serine protease pepD |

|
 0.30163 0.30163
| |
| Rv1762c | hypothetical protein |

|
 0.30093 0.30093
| |
| Rv0138 | conserved hypothetical protein |

|
 0.30066 0.30066
|