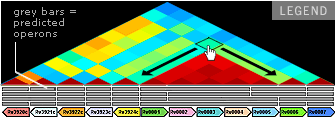
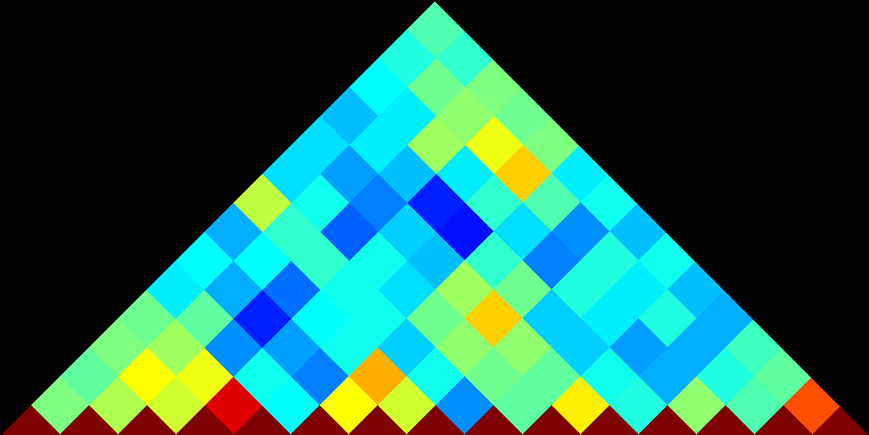
OPERON CORRELATION BROWSER
Explore how genes influence each other’s expression. Each diamond shape represents the correlation in expression for the two genes located at the end of the extended diagonals. Dark red represents the strongest positive correlation. Mouse over the diamonds to see the Pearson correlation coefficient for each gene pair.