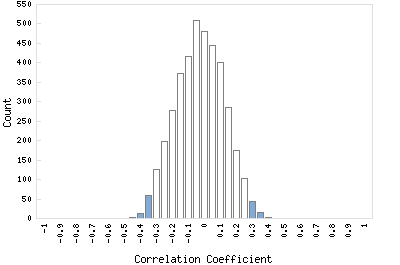
Correlation Catalog M. tuberculosis H37Rv: Rv3764c - two component system sensor kinase
Distribution of gene correlation: Rv3764c vs. all genes
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0951 | succinyl-CoA synthetase beta chain sucC |

|
 -0.43583 -0.43583
| |
| Rv1523 | methyltransferase |


|
 -0.43534 -0.43534
| |
| Rv3213c | chromosome partitioning protein |

|
 -0.42006 -0.42006
| |
| Rv3099c | conserved hypothetical protein |

|
 -0.38133 -0.38133
| |
| Rv0925c | conserved hypothetical protein |

|
 -0.37326 -0.37326
| |
| Rv0723 | 50S ribosomal protein L15 rplO |

|
 -0.36974 -0.36974
| |
| Rv3031 | conserved hypothetical protein |

|
 -0.36974 -0.36974
| |
| Rv2289 | cdp-diacylglycerol pyrophosphatase cdh |

|
 -0.36931 -0.36931
| |
| Rv0199 | conserved membrane protein |

|
 -0.36797 -0.36797
| |
| Rv0462 | dihydrolipoamide dehydrogenase lpd |

|
 -0.35989 -0.35989
| |
| Rv0652 | 50S ribosomal protein L7/L12 rplL |

|
 -0.35918 -0.35918
| |
| Rv2213 | aminopeptidase pepB |

|
 -0.35725 -0.35725
| |
| Rv0761c | zinc-type alcohol dehydrogenase NAD dependent adhB |

|
 -0.35692 -0.35692
| |
| Rv0009 | iron-regulated peptidyl-prolyl-cis-trans-isomerase A ppiA |

|
 -0.35524 -0.35524
| |
| Rv1175c | NADPH dependent 2,4-dienoyl-CoA reductase fadH |

|
 -0.35401 -0.35401
| |
| Rv0432 | periplasmic superoxide dismutase sodC |

|
 -0.35329 -0.35329
| |
| Rv3747 | conserved hypothetical protein |

|
 -0.34716 -0.34716
| |
| Rv3011c | glutamyl-tRNA(gln) amidotransferase subunit A gatA |

|
 -0.34522 -0.34522
| |
| Rv3679 | anion transporter ATPase |

|
 -0.34345 -0.34345
| |
| Rv0905 | enoyl-CoA hydratase echA6 |

|
 -0.34176 -0.34176
| |
| Rv1303 | conserved membrane protein |

|
 -0.34082 -0.34082
| |
| Rv2069 | RNA polymerase sigma factor sigC |

|
 -0.33887 -0.33887
| |
| Rv0440 | 60 kda chaperonin 2 groEL2 |

|
 -0.33858 -0.33858
| |
| Rv2889c | elongation factor tsf |

|
 -0.3368 -0.3368
| |
| Rv1111c | conserved hypothetical protein |

|
 -0.33678 -0.33678
| |
| Rv2908c | conserved hypothetical protein |

|
 -0.33521 -0.33521
| |
| Rv0957 | bifunctional purine biosynthesis protein purH |

|
 -0.33499 -0.33499
| |
| Rv2845c | prolyl-tRNA synthetase proS |

|
 -0.33388 -0.33388
| |
| Rv2561 | conserved hypothetical protein |

|
 -0.33254 -0.33254
| |
| Rv0418 | lipoprotein aminopeptidase lpqL |

|
 -0.33232 -0.33232
| |
| Rv2843 | conserved alanine rich membrane protein |

|
 -0.33165 -0.33165
| |
| Rv3119 | molybdenum cofactor biosynthesis protein E moaE1 |

|
 -0.33115 -0.33115
| |
| Rv0719 | 50S ribosomal protein L6 rplF |

|
 -0.33045 -0.33045
| |
| Rv3417c | 60 kda chaperonin 1 groEL1 |

|
 -0.3302 -0.3302
| |
| Rv1611 | indole-3-glycerol phosphate synthase trpC |

|
 -0.32915 -0.32915
| |
| Rv1241 | conserved hypothetical protein |

|
 -0.32882 -0.32882
| |
| Rv3224 | iron-regulated short-chain dehydrogenase/reductase |


|
 -0.32433 -0.32433
| |
| Rv0248c | succinate dehydrogenase iron-sulfur subunit |

|
 -0.32374 -0.32374
| |
| Rv1071c | enoyl-CoA hydratase echA9 |

|
 -0.32316 -0.32316
| |
| Rv1590 | conserved hypothetical protein |

|
 -0.32174 -0.32174
| |
| Rv2194 | ubiquinol-cytochrome C reductase qcrC cytochrome C subunit |

|
 -0.3205 -0.3205
| |
| Rv3803c | secreted fibronectin-binding protein antigen protein fbpD |

|
 -0.32003 -0.32003
| |
| Rv1780 | conserved hypothetical protein |

|
 -0.31912 -0.31912
| |
| Rv2329c | nitrite extrusion protein 1 narK1 |

|
 -0.31888 -0.31888
| |
| Rv2981c | D-alanine-D-alanine ligase ddlA |

|
 -0.31881 -0.31881
| |
| Rv3465 | dTDP-4-dehydrorhamnose 3,5-epimerase rmlC |

|
 -0.31878 -0.31878
| |
| Rv1227c | transmembrane protein |

|
 -0.31789 -0.31789
| |
| Rv2191 | conserved hypothetical protein |

|
 -0.31542 -0.31542
| |
| Rv0533c | 3-oxoacyl-[acyl-carrier-protein] synthase III fabH |

|
 -0.31529 -0.31529
| |
| Rv1860 | alanine and proline rich secreted protein apa |

|
 -0.31486 -0.31486
| |
| Rv0947c | mycolyl transferase |

|
 -0.31408 -0.31408
| |
| Rv2888c | amidase amiC |

|
 -0.31401 -0.31401
| |
| Rv1932 | thiol peroxidase tpx |

|
 -0.31139 -0.31139
| |
| Rv1436 | glyceraldehyde 3-phosphate dehydrogenase gap |

|
 -0.31061 -0.31061
| |
| Rv0545c | low affinity inorganic phosphate transporter membrane protein pitA |

|
 -0.3096 -0.3096
| |
| Rv2890c | 30S ribosomal protein S2 rpsB |

|
 -0.30816 -0.30816
| |
| Rv1428c | conserved hypothetical protein |

|
 -0.30709 -0.30709
| |
| Rv1305 | ATP synthase C chain atpE |

|
 -0.30674 -0.30674
| |
| Rv0583c | lipoprotein lpqN |

|
 -0.3066 -0.3066
| |
| Rv0708 | 50S ribosomal protein L16 rplP |

|
 -0.30619 -0.30619
| |
| Rv0651 | 50S ribosomal protein L10 rplJ |

|
 -0.30574 -0.30574
| |
| Rv0429c | polypeptide deformylase def |

|
 -0.30529 -0.30529
| |
| Rv2450c | resuscitation-promoting factor rpfE |

|
 -0.30391 -0.30391
| |
| Rv1191 | conserved hypothetical protein |

|
 -0.3038 -0.3038
| |
| Rv0713 | conserved membrane protein |

|
 -0.30367 -0.30367
| |
| Rv2909c | 30S ribosomal protein S16 rpsP |

|
 -0.30354 -0.30354
| |
| Rv2113 | membrane protein |

|
 -0.3031 -0.3031
| |
| Rv2357c | glycyl-tRNA synthetase glyS |

|
 -0.30292 -0.30292
| |
| Rv1133c | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase metE |

|
 -0.30272 -0.30272
| |
| Rv1211 | conserved hypothetical protein |

|
 -0.30268 -0.30268
| |
| Rv0460 | conserved hydrophobic protein |

|
 -0.30202 -0.30202
| |
| Rv2768c | PPE family protein |

|
 -0.30183 -0.30183
| |
| Rv0249c | succinate dehydrogenase membrane anchor subunit |

|
 -0.30141 -0.30141
| |
| Rv1311 | ATP synthase epsilon chain atpC |

|
 -0.30008 -0.30008
| |
| Rv2171 | lipoprotein lppM |

|
 -0.30002 -0.30002
|
Genes with positive correlation 
Categories Enriched  : O
: O
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0983 | serine protease pepD |

|
 0.43959 0.43959
| |
| Rv3446c | hypothetical alanine and valine rich protein |

|
 0.42407 0.42407
| |
| Rv2270 | lipoprotein lppN |

|
 0.41275 0.41275
| |
| Rv3706c | conserved proline rich protein |

|
 0.39998 0.39998
| |
| Rv3830c | transcriptional regulator, tetR-family |

|
 0.38949 0.38949
| |
| Rv2976c | uracil-DNA glycosylase ung |

|
 0.38837 0.38837
| |
| Rv3366 | tRNA/rRNA methylase spoU |

|
 0.38536 0.38536
| |
| Rv3064c | conserved membrane protein |

|
 0.38364 0.38364
| |
| Rv1126c | conserved hypothetical protein |

|
 0.37047 0.37047
| |
| Rv0991c | conserved serine rich protein |

|
 0.36124 0.36124
| |
| Rv0690c | conserved hypothetical protein |

|
 0.35721 0.35721
| |
| Rv1868 | conserved hypothetical protein |


|
 0.35703 0.35703
| |
| Rv3618 | monooxygenase |

|
 0.3569 0.3569
| |
| Rv1862 | alcohol dehydrogenase adhA |

|
 0.35648 0.35648
| |
| Rv3834c | seryl-tRNA synthetase serS |

|
 0.35628 0.35628
| |
| Rv0982 | two component system sensor kinase mprB |

|
 0.35536 0.35536
| |
| Rv3730c | conserved hypothetical protein |

|
 0.35085 0.35085
| |
| Rv2262c | conserved hypothetical protein |

|
 0.35014 0.35014
| |
| Rv0870c | conserved membrane protein |

|
 0.34916 0.34916
| |
| Rv1118c | conserved hypothetical protein |

|
 0.34809 0.34809
| |
| Rv0724 | protease IV sppA |


|
 0.34468 0.34468
| |
| Rv0758 | two component system sensor kinase phoR |

|
 0.34097 0.34097
| |
| Rv3758c | osmoprotectant proV |

|
 0.33702 0.33702
| |
| Rv2711 | iron-dependent repressor and activator ideR |

|
 0.33314 0.33314
| |
| Rv3238c | conserved membrane protein |

|
 0.33036 0.33036
| |
| Rv3394c | conserved hypothetical protein |

|
 0.32997 0.32997
| |
| Rv1846c | transcriptional regulator |

|
 0.32909 0.32909
| |
| Rv1720c | conserved hypothetical protein |

|
 0.32863 0.32863
| |
| Rv2800 | hydrolase |

|
 0.32648 0.32648
| |
| Rv3070 | conserved membrane protein |

|
 0.32604 0.32604
| |
| Rv0738 | conserved hypothetical protein |

|
 0.3246 0.3246
| |
| Rv0262c | aminoglycoside 2-N-acetyltransferase aac |

|
 0.32014 0.32014
| |
| Rv0568 | cytochrome P450 135B1 cyp135B1 |

|
 0.31939 0.31939
| |
| Rv3386 | transposase |

|
 0.31864 0.31864
| |
| Rv3802c | conserved membrane protein |

|
 0.31821 0.31821
| |
| Rv2037c | conserved membrane protein |

|
 0.31678 0.31678
| |
| Rv2052c | conserved hypothetical protein |

|
 0.31633 0.31633
| |
| Rv0967 | conserved hypothetical protein |

|
 0.31576 0.31576
| |
| Rv0981 | mycobacterial persistence regulator mprA |


|
 0.31507 0.31507
| |
| Rv0356c | conserved hypothetical protein |

|
 0.31486 0.31486
| |
| Rv3866 | conserved hypothetical protein |

|
 0.31319 0.31319
| |
| Rv3046c | conserved hypothetical protein |

|
 0.31265 0.31265
| |
| Rv0978c | PE-PGRS family protein |

|
 0.3124 0.3124
| |
| Rv3785 | hypothetical protein |

|
 0.31124 0.31124
| |
| Rv2710 | RNA polymerase sigma factor sigB |

|
 0.31085 0.31085
| |
| Rv1530 | alcohol dehydrogenase adh |


|
 0.31053 0.31053
| |
| Rv2224c | exported protease |

|
 0.31025 0.31025
| |
| Rv0376c | conserved hypothetical protein |

|
 0.30924 0.30924
| |
| Rv1072 | conserved membrane protein |

|
 0.30874 0.30874
| |
| Rv3734c | conserved hypothetical protein |

|
 0.30837 0.30837
| |
| Rv2110c | proteasome beta subunit prcB |

|
 0.30779 0.30779
| |
| Rv1362c | membrane protein |

|
 0.30769 0.30769
| |
| Rv2616 | conserved hypothetical protein |

|
 0.30755 0.30755
| |
| Rv0980c | PE-PGRS family protein |

|
 0.30549 0.30549
| |
| Rv0522 | gabA permease gabP |

|
 0.30431 0.30431
| |
| Rv2042c | conserved hypothetical protein |

|
 0.30382 0.30382
| |
| Rv0877 | conserved hypothetical protein |

|
 0.30368 0.30368
| |
| Rv2866 | conserved hypothetical protein |

|
 0.30246 0.30246
| |
| Rv2686c | antibiotic-transport membrane leucine and alanine and valine rich protein ABC transporter |

|
 0.30092 0.30092
| |
| Rv2830c | conserved hypothetical protein |

|
 0.30092 0.30092
| |
| Rv1002c | conserved membrane protein |

|
 0.30044 0.30044
|