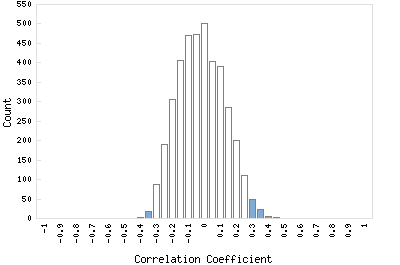
Correlation Catalog M. tuberculosis H37Rv: Rv3529c - conserved hypothetical protein
Distribution of gene correlation: Rv3529c vs. all genes
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv2184c | conserved hypothetical protein |

|
 -0.39555 -0.39555
| |
| Rv2585c | conserved lipoprotein |


|
 -0.35612 -0.35612
| |
| Rv1848 | urease gamma subunit ureA |

|
 -0.34779 -0.34779
| |
| Rv1002c | conserved membrane protein |

|
 -0.33359 -0.33359
| |
| Rv1060 | hypothetical protein |

|
 -0.33311 -0.33311
| |
| Rv0734 | methionine aminopeptidase mapA |

|
 -0.33237 -0.33237
| |
| Rv3376 | conserved hypothetical protein |

|
 -0.33055 -0.33055
| |
| Rv2532c | hypothetical protein |

|
 -0.32859 -0.32859
| |
| Rv3028c | electron transfer flavoprotein alpha subunit fixB |

|
 -0.32269 -0.32269
| |
| Rv3648c | cold shock protein A cspA |

|
 -0.32185 -0.32185
| |
| Rv0434 | conserved hypothetical protein |

|
 -0.31258 -0.31258
| |
| Rv0198c | zinc metalloprotease |

|
 -0.31172 -0.31172
| |
| Rv3461c | 50S ribosomal protein L36 rpmJ |

|
 -0.3107 -0.3107
| |
| Rv3460c | 30S ribosomal protein S13 rpsM |

|
 -0.3099 -0.3099
| |
| Rv0954 | conserved membrane protein |

|
 -0.30935 -0.30935
| |
| Rv2992c | glutamyl-tRNA synthetase gltS |

|
 -0.30739 -0.30739
| |
| Rv1097c | glycine and proline rich membrane protein |

|
 -0.30561 -0.30561
| |
| Rv1626 | two component system transcriptional regulator |

|
 -0.30558 -0.30558
| |
| Rv2144c | transmembrane protein |

|
 -0.30525 -0.30525
| |
| Rv0640 | 50S ribosomal protein L11 rplK |

|
 -0.30127 -0.30127
|
Genes with positive correlation 
Categories Enriched  : I
: I
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv3531c | hypothetical protein |

|
 0.55638 0.55638
| |
| Rv3544c | acyl-CoA dehydrogenase fadE28 |

|
 0.5133 0.5133
| |
| Rv3543c | acyl-CoA dehydrogenase fadE29 |

|
 0.48105 0.48105
| |
| Rv3530c | oxidoreductase |


|
 0.47578 0.47578
| |
| Rv3538 | dehydrogenase |

|
 0.45994 0.45994
| |
| Rv1939 | oxidoreductase |

|
 0.42696 0.42696
| |
| Rv3545c | cytochrome P450 125 cyp125 |

|
 0.40901 0.40901
| |
| Rv3537 | dehydrogenase |

|
 0.4024 0.4024
| |
| Rv1777 | cytochrome P450 144 cyp144 |

|
 0.40023 0.40023
| |
| Rv3541c | conserved hypothetical protein |

|
 0.38907 0.38907
| |
| Rv3314c | thymidine phosphorylase deoA |

|
 0.38591 0.38591
| |
| Rv3553 | oxidoreductase |

|
 0.38379 0.38379
| |
| Rv3359 | oxidoreductase |

|
 0.3788 0.3788
| |
| Rv3550 | enoyl-CoA hydratase echA20 |

|
 0.37814 0.37814
| |
| Rv3527 | hypothetical protein |

|
 0.37759 0.37759
| |
| Rv1949c | conserved hypothetical protein |

|
 0.37756 0.37756
| |
| Rv0897c | oxidoreductase |

|
 0.37588 0.37588
| |
| Rv3567c | oxidoreductase |

|
 0.37362 0.37362
| |
| Rv2043c | pyrazinamidase/nicotinamidas pncA |

|
 0.37253 0.37253
| |
| Rv1354c | conserved hypothetical protein |

|
 0.37052 0.37052
| |
| Rv2577 | conserved hypothetical protein |

|
 0.37047 0.37047
| |
| Rv3535c | acetaldehyde dehydrogenase |

|
 0.36933 0.36933
| |
| Rv3568c | biphenyl-2,3-diol 1,2-dioxygenase bphC |

|
 0.36288 0.36288
| |
| Rv2551c | conserved hypothetical protein |

|
 0.36076 0.36076
| |
| Rv3515c | fatty-acid-CoA ligase fadD19 |


|
 0.3601 0.3601
| |
| Rv2317 | sugar-transport membrane protein ABC transporter uspB |

|
 0.35937 0.35937
| |
| Rv3289c | transmembrane protein |

|
 0.35924 0.35924
| |
| Rv1969 | MCE-family protein mce3D |

|
 0.35841 0.35841
| |
| Rv1931c | transcriptional regulator |

|
 0.3547 0.3547
| |
| Rv1537 | DNA polymerase IV dinX |

|
 0.35404 0.35404
| |
| Rv0223c | aldehyde dehydrogenase |

|
 0.3524 0.3524
| |
| Rv1029 | potassium-transporting ATPase A chain kdpA |

|
 0.34976 0.34976
| |
| Rv2437 | conserved hypothetical protein |

|
 0.34794 0.34794
| |
| Rv3097c | PE-PGRS family protein |

|
 0.34387 0.34387
| |
| Rv3505 | acyl-CoA dehydrogenase fadE27 |

|
 0.34078 0.34078
| |
| Rv0441c | hypothetical protein |

|
 0.34048 0.34048
| |
| Rv3809c | UDP-galactopyranose mutase glf |

|
 0.33675 0.33675
| |
| Rv3437 | conserved membrane protein |

|
 0.3362 0.3362
| |
| Rv0812 | amino acid aminotransferase |


|
 0.33106 0.33106
| |
| Rv3871 | conserved hypothetical protein |

|
 0.32801 0.32801
| |
| Rv2233 |

|
 0.32644 0.32644
| ||
| Rv3079c | conserved hypothetical protein |

|
 0.32447 0.32447
| |
| Rv3347c | PPE family protein |

|
 0.32313 0.32313
| |
| Rv2359 | ferric uptake regulation protein furB |

|
 0.32286 0.32286
| |
| Rv2378c | lysine-N-oxygenase mbtG |

|
 0.32255 0.32255
| |
| Rv3546 | acetyl-CoA acetyltransferase fadA5 |

|
 0.32253 0.32253
| |
| Rv1161 | respiratory nitrate reductase alpha chain narG |

|
 0.32251 0.32251
| |
| Rv2384 | bifunctional salicyl-AMP ligase/salicyl-S-arcp synthetase mbtA |

|
 0.32142 0.32142
| |
| Rv3523 | lipid carrier protein or keto acyl-CoA thiolase ltp3 |

|
 0.32008 0.32008
| |
| Rv3563 | acyl-CoA dehydrogenase fadE32 |

|
 0.31672 0.31672
| |
| Rv1231c | membrane protein |

|
 0.31542 0.31542
| |
| Rv2003c | conserved hypothetical protein |


|
 0.31358 0.31358
| |
| Rv3349c | transposase |

|
 0.31301 0.31301
| |
| Rv2328 | PE family protein |

|
 0.31282 0.31282
| |
| Rv0591 | MCE-family protein mce2C |

|
 0.31263 0.31263
| |
| Rv0397 | 13E12 repeat family protein |

|
 0.31223 0.31223
| |
| Rv1499 | hypothetical protein |

|
 0.31222 0.31222
| |
| Rv1748 | hypothetical protein |

|
 0.31117 0.31117
| |
| Rv2706c | hypothetical protein |

|
 0.31091 0.31091
| |
| Rv0573c | conserved hypothetical protein |

|
 0.30976 0.30976
| |
| Rv2338c | molybdopterin biosynthesis protein moeW |

|
 0.30929 0.30929
| |
| Rv2170 | conserved hypothetical protein |

|
 0.30918 0.30918
| |
| Rv3755c | conserved hypothetical protein |

|
 0.30915 0.30915
| |
| Rv2042c | conserved hypothetical protein |

|
 0.30842 0.30842
| |
| Rv3526 | oxidoreductase |


|
 0.30834 0.30834
| |
| Rv0149 | quinone oxidoreductase |


|
 0.30797 0.30797
| |
| Rv1131 | citrate synthase I gltA1 |

|
 0.30767 0.30767
| |
| Rv0311 | hypothetical protein |

|
 0.30766 0.30766
| |
| Rv2679 | enoyl-CoA hydratase echA15 |

|
 0.30713 0.30713
| |
| Rv1985c | transcriptional regulator, lysR-family |

|
 0.30712 0.30712
| |
| Rv3242c | conserved hypothetical protein |

|
 0.30585 0.30585
| |
| Rv3552 | CoA-transferase beta subunit |

|
 0.30584 0.30584
| |
| Rv3333c | hypothetical proline rich protein |

|
 0.30533 0.30533
| |
| Rv1291c | conserved secreted protein |

|
 0.3041 0.3041
| |
| Rv2330c | lipoprotein lppP |

|
 0.30264 0.30264
| |
| Rv1163 | respiratory nitrate reductase delta chain narJ |

|
 0.30227 0.30227
| |
| Rv3023c | transposase |

|
 0.30212 0.30212
| |
| Rv2385 | acetyl hydrolase mbtJ |

|
 0.30148 0.30148
| |
| Rv3387 | transposase |

|
 0.30141 0.30141
| |
| Rv2558 | conserved hypothetical protein |

|
 0.30131 0.30131
| |
| Rv3290c | L-lysine-epsilon aminotransferase lat |

|
 0.30053 0.30053
|