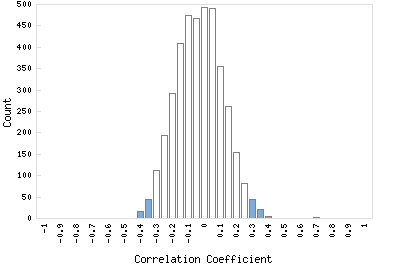
Correlation Catalog M. tuberculosis H37Rv: Rv3323c - moaD-moaE fusion protein moaX
Distribution of gene correlation: Rv3323c vs. all genes
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0467 | isocitrate lyase icl |

|
 -0.40321 -0.40321
| |
| Rv1778c | hypothetical protein |

|
 -0.39503 -0.39503
| |
| Rv2172c | conserved hypothetical protein |

|
 -0.39408 -0.39408
| |
| Rv2931 | phenolpthiocerol synthesis type-I polyketide synthase ppsA |

|
 -0.38219 -0.38219
| |
| Rv2542 | conserved hypothetical protein |

|
 -0.38064 -0.38064
| |
| Rv2930 | fatty-acid-CoA ligase fadD26 |


|
 -0.37877 -0.37877
| |
| Rv2940c | multifunctional mycocerosic acid synthase membrane-associated mas |

|
 -0.37168 -0.37168
| |
| Rv1483 | 3-oxoacyl-[acyl-carrier protein] reductase fabG1 |


|
 -0.37089 -0.37089
| |
| Rv1789 | PPE family protein |

|
 -0.36601 -0.36601
| |
| Rv0485 | transcriptional regulator |

|
 -0.36483 -0.36483
| |
| Rv2837c | conserved hypothetical protein |

|
 -0.3638 -0.3638
| |
| Rv2933 | phenolpthiocerol synthesis type-I polyketide synthase ppsC |

|
 -0.36355 -0.36355
| |
| Rv3093c | oxidoreductase |

|
 -0.36149 -0.36149
| |
| Rv1779c | hypothetical membrane protein |

|
 -0.36098 -0.36098
| |
| Rv1143 | alpha-methylacyl-CoA racemase mcr |

|
 -0.36073 -0.36073
| |
| Rv2640c | transcriptional regulator, arsR-family |

|
 -0.35973 -0.35973
| |
| Rv1127c | pyruvate, phosphate dikinase ppdK |

|
 -0.35339 -0.35339
| |
| Rv2935 | phenolpthiocerol synthesis type-I polyketide synthase ppsE |

|
 -0.34709 -0.34709
| |
| Rv3013 | conserved hypothetical protein |

|
 -0.34694 -0.34694
| |
| Rv0476 | conserved membrane protein |

|
 -0.34319 -0.34319
| |
| Rv0973c | acetyl-/propionyl-CoA carboxylase alpha subunit accA2 |

|
 -0.34169 -0.34169
| |
| Rv3003c | acetolactate synthase large subunit ilvB1 |
![COG0028: (EH) Thiamine pyrophosphate-requiring enzymes [acetolactate synthase; pyruvate dehydrogenase (cytochrome); glyoxylate carboligase; phosphonopyruvate decarboxylase]](supportFiles/E.png)
![COG0028: (EH) Thiamine pyrophosphate-requiring enzymes [acetolactate synthase; pyruvate dehydrogenase (cytochrome); glyoxylate carboligase; phosphonopyruvate decarboxylase]](supportFiles/H.png)
|
 -0.33771 -0.33771
| |
| Rv1485 | ferrochelatase hemZ |

|
 -0.33563 -0.33563
| |
| Rv0483 | lipoprotein lprQ |

|
 -0.33561 -0.33561
| |
| Rv2937 | daunorubicin-dim-transport membrane protein ABC transporter drrB |

|
 -0.33495 -0.33495
| |
| Rv2704 | conserved hypothetical protein |

|
 -0.33281 -0.33281
| |
| Rv1361c | PPE family protein |

|
 -0.33187 -0.33187
| |
| Rv3137 | monophosphatase |

|
 -0.33177 -0.33177
| |
| Rv1009 | resuscitation-promoting factor rpfB |

|
 -0.33175 -0.33175
| |
| Rv2959c | methyltransferase/methylase |

|
 -0.32955 -0.32955
| |
| Rv2329c | nitrite extrusion protein 1 narK1 |

|
 -0.32934 -0.32934
| |
| Rv2932 | phenolpthiocerol synthesis type-I polyketide synthase ppsB |

|
 -0.32701 -0.32701
| |
| Rv3001c | ketol-acid reductoisomerase ilvC |


|
 -0.3259 -0.3259
| |
| Rv0466 | conserved hypothetical protein |

|
 -0.32536 -0.32536
| |
| Rv0327c | cytochrome P450 135A1 cyp135A1 |

|
 -0.32403 -0.32403
| |
| Rv0860 | fatty-acid oxidation protein fadB |

|
 -0.32274 -0.32274
| |
| Rv0390 | conserved hypothetical protein |

|
 -0.32149 -0.32149
| |
| Rv3455c | tRNA pseudouridine synthase A truA |

|
 -0.3212 -0.3212
| |
| Rv2645 | hypothetical protein |

|
 -0.32112 -0.32112
| |
| Rv0720 | 50S ribosomal protein L18 rplR |

|
 -0.3207 -0.3207
| |
| Rv2947c | polyketide synthase pks15 |

|
 -0.31679 -0.31679
| |
| Rv3667 | acetyl-CoA synthetase acs |

|
 -0.3165 -0.3165
| |
| Rv0905 | enoyl-CoA hydratase echA6 |

|
 -0.3164 -0.3164
| |
| Rv2697c | deoxyuridine 5-triphosphate nucleotidohydrolase dut |

|
 -0.31532 -0.31532
| |
| Rv0469 | mycolic acid synthase umaA |

|
 -0.31442 -0.31442
| |
| Rv2429 | alkyl hydroperoxide reductase D protein ahpD |

|
 -0.31385 -0.31385
| |
| Rv1381 | dihydroorotase pyrC |

|
 -0.3135 -0.3135
| |
| Rv3737 | conserved membrane protein |

|
 -0.31214 -0.31214
| |
| Rv2327 | conserved hypothetical protein |

|
 -0.31089 -0.31089
| |
| Rv2838c | ribosome-binding factor A rbfA |

|
 -0.31086 -0.31086
| |
| Rv0646c | lipase/esterase lipG |

|
 -0.30979 -0.30979
| |
| Rv2450c | resuscitation-promoting factor rpfE |

|
 -0.30964 -0.30964
| |
| Rv1420 | excinuclease ABC subunit C uvrC |

|
 -0.30925 -0.30925
| |
| Rv2421c | nicotinate-nucleotide adenylyltransferase nadD |

|
 -0.30892 -0.30892
| |
| Rv2431c | PE family protein |

|
 -0.30872 -0.30872
| |
| Rv0475 | iron-regulated heparin binding hemagglutinin hbhA |

|
 -0.30656 -0.30656
| |
| Rv3804c | secreted fibronectin-binding protein antigen fbpA |

|
 -0.30587 -0.30587
| |
| Rv3167c | transcriptional regulator, tetR-family |

|
 -0.30573 -0.30573
| |
| Rv1096 | glycosyl hydrolase |

|
 -0.30542 -0.30542
| |
| Rv3135 | PPE family protein |

|
 -0.30403 -0.30403
| |
| Rv2417c | conserved hypothetical protein |

|
 -0.30152 -0.30152
| |
| Rv1627c | nonspecific lipid-transfer protein |

|
 -0.3008 -0.3008
|
Genes with positive correlation 
Categories Enriched  : C
: C
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv3324c | molybdenum cofactor biosynthesis protein C 3 moaC3 |

|
 0.74555 0.74555
| |
| Rv3322c | methyltransferase |


|
 0.71918 0.71918
| |
| Rv3321c | conserved hypothetical protein |

|
 0.57656 0.57656
| |
| Rv3144c | PPE family protein |

|
 0.54184 0.54184
| |
| Rv3158 | NADH dehydrogenase I chain N nuoN |

|
 0.46292 0.46292
| |
| Rv3156 | NADH dehydrogenase I chain L nuoL |


|
 0.4466 0.4466
| |
| Rv1881c | lipoprotein lppE |

|
 0.41818 0.41818
| |
| Rv0060 | conserved hypothetical protein |

|
 0.4078 0.4078
| |
| Rv1497 | esterase lipL |

|
 0.40353 0.40353
| |
| Rv3146 | NADH dehydrogenase I chain B nuoB |

|
 0.39941 0.39941
| |
| Rv1754c | conserved hypothetical protein |

|
 0.39792 0.39792
| |
| Rv0276 | conserved hypothetical protein |

|
 0.39483 0.39483
| |
| Rv3164c | methanol dehydrogenase transcriptional regulator moxR3 |

|
 0.39074 0.39074
| |
| Rv3142c | hypothetical protein |

|
 0.38257 0.38257
| |
| Rv2632c | conserved hypothetical protein |

|
 0.38057 0.38057
| |
| Rv1906c | conserved hypothetical protein |

|
 0.38048 0.38048
| |
| Rv0995 | ribosomal-protein-alanine acetyltransferase rimJ |

|
 0.37597 0.37597
| |
| Rv3295 | transcriptional regulator, tetR-family |

|
 0.37371 0.37371
| |
| Rv1478 | invasion-associated protein |

|
 0.37214 0.37214
| |
| Rv1882c | short-chain type dehydrogenase/reductase |


|
 0.37089 0.37089
| |
| Rv0272c | hypothetical protein |

|
 0.36767 0.36767
| |
| Rv3152 | NADH dehydrogenase I chain H nuoH |

|
 0.36027 0.36027
| |
| Rv0166 | fatty-acid-CoA ligase fadD5 |


|
 0.35801 0.35801
| |
| Rv0108c | hypothetical protein |

|
 0.35797 0.35797
| |
| Rv3148 | NADH dehydrogenase I chain D nuoD |

|
 0.35629 0.35629
| |
| Rv0174 | MCE-family protein mce1F |

|
 0.35358 0.35358
| |
| Rv3312c | conserved hypothetical protein |

|
 0.35328 0.35328
| |
| Rv0655 | ribonucleotide-transport ATP-binding protein ABC transporter mkl |

|
 0.35275 0.35275
| |
| Rv1444c | hypothetical protein |

|
 0.352 0.352
| |
| Rv3296 | ATP-dependent helicase lhr |

|
 0.35047 0.35047
| |
| Rv0168 | hypothetical membrane protein yrbE1B |

|
 0.35027 0.35027
| |
| Rv2722 | conserved hypothetical protein |

|
 0.34663 0.34663
| |
| Rv3151 | NADH dehydrogenase I chain G nuoG |

|
 0.34636 0.34636
| |
| Rv0679c | conserved threonine rich protein |

|
 0.34337 0.34337
| |
| Rv3147 | NADH dehydrogenase I chain C nuoC |

|
 0.3427 0.3427
| |
| Rv1829 | conserved hypothetical protein |

|
 0.34058 0.34058
| |
| Rv3583c | transcriptional regulator |

|
 0.34007 0.34007
| |
| Rv3058c | transcriptional regulator, tetR-family |

|
 0.3387 0.3387
| |
| Rv2092c | ATP-dependent DNA helicase helY |



|
 0.33751 0.33751
| |
| Rv3083 | monooxygenase |

|
 0.33542 0.33542
| |
| Rv0173 | MCE-family lipoprotein mce1E |

|
 0.33492 0.33492
| |
| Rv3241c | conserved hypothetical protein |

|
 0.33288 0.33288
| |
| Rv3633 | conserved hypothetical protein |

|
 0.33095 0.33095
| |
| Rv3168 | conserved hypothetical protein |

|
 0.33058 0.33058
| |
| Rv3089 | fatty-acid-CoA ligase fadD13 |


|
 0.32939 0.32939
| |
| Rv1662 | polyketide synthase pks8 |

|
 0.32912 0.32912
| |
| Rv3244c | lipoprotein lpqB |

|
 0.32826 0.32826
| |
| Rv2409c | conserved hypothetical protein |

|
 0.32571 0.32571
| |
| Rv0755c | PPE family protein |

|
 0.32427 0.32427
| |
| Rv0111 | transmembrane acyltransferase |

|
 0.32412 0.32412
| |
| Rv0868c | molybdenum cofactor biosynthesis protein D 2 moaD2 |

|
 0.3241 0.3241
| |
| Rv3087 | conserved hypothetical protein |

|
 0.32356 0.32356
| |
| Rv3059 | cytochrome P450 136 cyp136 |

|
 0.32143 0.32143
| |
| Rv2484c | conserved hypothetical protein |

|
 0.31999 0.31999
| |
| Rv3209 | conserved proline and threonine rich protein |

|
 0.31953 0.31953
| |
| Rv3313c | adenosine deaminase add |

|
 0.31774 0.31774
| |
| Rv3149 | NADH dehydrogenase I chain E nuoE |

|
 0.31717 0.31717
| |
| Rv3154 | NADH dehydrogenase I chain J nuoJ |

|
 0.3171 0.3171
| |
| Rv0221 | conserved hypothetical protein |

|
 0.31578 0.31578
| |
| Rv3272 | conserved hypothetical protein |

|
 0.31539 0.31539
| |
| Rv3150 | NADH dehydrogenase I chain F nuoF |

|
 0.31502 0.31502
| |
| Rv0172 | MCE-family protein mce1D |

|
 0.3129 0.3129
| |
| Rv0314c | conserved membrane protein |

|
 0.31201 0.31201
| |
| Rv3642c | hypothetical protein |

|
 0.31095 0.31095
| |
| Rv3734c | conserved hypothetical protein |

|
 0.30989 0.30989
| |
| Rv0074 | conserved hypothetical protein |

|
 0.30971 0.30971
| |
| Rv3155 | NADH dehydrogenase I chain K nuoK |

|
 0.30827 0.30827
| |
| Rv0239 | conserved hypothetical protein |

|
 0.30489 0.30489
| |
| Rv1770 | conserved hypothetical protein |

|
 0.30436 0.30436
| |
| Rv2750 | dehydrogenase |


|
 0.30373 0.30373
| |
| Rv3491 | hypothetical protein |

|
 0.30344 0.30344
| |
| Rv0680c | conserved membrane protein |

|
 0.30267 0.30267
| |
| Rv0208c | methlytransferase |

|
 0.30194 0.30194
| |
| Rv0170 | MCE-family protein mce1B |

|
 0.30122 0.30122
| |
| Rv2507 | conserved proline rich membrane protein |

|
 0.30051 0.30051
| |
| Rv3190c | hypothetical protein |

|
 0.30017 0.30017
|