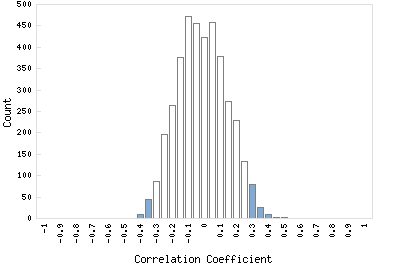
Correlation Catalog M. tuberculosis H37Rv: Rv2671 - bifunctional riboflavin biosynthesis protein ribD
Distribution of gene correlation: Rv2671 vs. all genes
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0299 | hypothetical protein |

|
 -0.41039 -0.41039
| |
| Rv1795 | conserved membrane protein |

|
 -0.37925 -0.37925
| |
| Rv1341 | conserved hypothetical protein |

|
 -0.37473 -0.37473
| |
| Rv3240c | preprotein translocase secA1 |

|
 -0.36693 -0.36693
| |
| Rv2861c | methionine aminopeptidase mapB |

|
 -0.36411 -0.36411
| |
| Rv3609c | GTP cyclohydrolase I folE |

|
 -0.35818 -0.35818
| |
| Rv2134c | conserved hypothetical protein |

|
 -0.35796 -0.35796
| |
| Rv0472c | transcriptional regulator, tetR-family |

|
 -0.35787 -0.35787
| |
| Rv0954 | conserved membrane protein |

|
 -0.35766 -0.35766
| |
| Rv1794 | conserved hypothetical protein |

|
 -0.35396 -0.35396
| |
| Rv1340 | ribonuclease rphA |

|
 -0.35364 -0.35364
| |
| Rv2051c | polyprenol-monophosphomannose synthase ppm1 |

|
 -0.34836 -0.34836
| |
| Rv3757c | osmoprotectant proW |

|
 -0.34691 -0.34691
| |
| Rv3028c | electron transfer flavoprotein alpha subunit fixB |

|
 -0.34365 -0.34365
| |
| Rv3117 | thiosulfate sulfurtransferase cysA3 |

|
 -0.34112 -0.34112
| |
| Rv0613c | hypothetical protein |

|
 -0.338 -0.338
| |
| Rv1792 | esat-6 like protein esxM |

|
 -0.33791 -0.33791
| |
| Rv2936 | daunorubicin-dim-transport ATP-binding protein ABC transporter drrA |

|
 -0.33637 -0.33637
| |
| Rv0722 | 50S ribosomal protein L30 rpmD |

|
 -0.33614 -0.33614
| |
| Rv2178c | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase aroG |

|
 -0.33572 -0.33572
| |
| Rv2164c | conserved proline rich membrane protein |

|
 -0.33174 -0.33174
| |
| Rv1843c | inosine-5-monophosphate dehydrogenase guaB1 |

|
 -0.32913 -0.32913
| |
| Rv1037c | Esat-6 like protein esxI |

|
 -0.32728 -0.32728
| |
| Rv0513 | conserved membrane protein |

|
 -0.32603 -0.32603
| |
| Rv1797 | conserved hypothetical protein |

|
 -0.32418 -0.32418
| |
| Rv0018c | serine/threonine phosphatase ppp |

|
 -0.32264 -0.32264
| |
| Rv1038c | Esat-6 like protein esxJ |

|
 -0.32068 -0.32068
| |
| Rv3257c | phosphomannomutase pmmA |

|
 -0.32028 -0.32028
| |
| Rv3024c | tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase trmU |

|
 -0.32023 -0.32023
| |
| Rv0157 | NAD(P) transhydrogenase beta subunit pntB |

|
 -0.31839 -0.31839
| |
| Rv1487 | conserved membrane protein |


|
 -0.31783 -0.31783
| |
| Rv3802c | conserved membrane protein |

|
 -0.3173 -0.3173
| |
| Rv1309 | ATP synthase gamma chain atpG |

|
 -0.31665 -0.31665
| |
| Rv0721 | 30S ribosomal protein S5 rpsE |

|
 -0.31578 -0.31578
| |
| Rv1328 | glycogen phosphorylase glgP |

|
 -0.31539 -0.31539
| |
| Rv2743c | conserved alanine rich membrane protein |

|
 -0.31324 -0.31324
| |
| Rv1310 | ATP synthase beta chain atpD |

|
 -0.31316 -0.31316
| |
| Rv3905c | Esat-6 like protein esxF |

|
 -0.31274 -0.31274
| |
| Rv3590c | PE-PGRS family protein |

|
 -0.31234 -0.31234
| |
| Rv0005 | DNA gyrase subunit B gyrB |

|
 -0.31196 -0.31196
| |
| Rv3003c | acetolactate synthase large subunit ilvB1 |
![COG0028: (EH) Thiamine pyrophosphate-requiring enzymes [acetolactate synthase; pyruvate dehydrogenase (cytochrome); glyoxylate carboligase; phosphonopyruvate decarboxylase]](supportFiles/E.png)
![COG0028: (EH) Thiamine pyrophosphate-requiring enzymes [acetolactate synthase; pyruvate dehydrogenase (cytochrome); glyoxylate carboligase; phosphonopyruvate decarboxylase]](supportFiles/H.png)
|
 -0.30976 -0.30976
| |
| Rv1297 | transcription termination factor rho |

|
 -0.30584 -0.30584
| |
| Rv2245 | 3-oxoacyl-[acyl-carrier protein] synthase 1 kasA |


|
 -0.3056 -0.3056
| |
| Rv0284 | conserved membrane protein |

|
 -0.30554 -0.30554
| |
| Rv2970c | lipase/esterase lipN |

|
 -0.30497 -0.30497
| |
| Rv1596 | nicotinate-nucleotide pyrophosphatase nadC |

|
 -0.30489 -0.30489
| |
| Rv1886c | secreted fibronectin-binding protein antigen 85-B fbpB |

|
 -0.30469 -0.30469
| |
| Rv0710 | 30S ribosomal protein S17 rpsQ |

|
 -0.30443 -0.30443
| |
| Rv2802c | hypothetical alanine and arginine rich protein |

|
 -0.30407 -0.30407
| |
| Rv0282 | conserved hypothetical protein |

|
 -0.30353 -0.30353
| |
| Rv0632c | enoyl-CoA hydratase echA3 |

|
 -0.30255 -0.30255
| |
| Rv3411c | inosine-5-monophosphate dehydrogenase guaB2 |

|
 -0.30063 -0.30063
| |
| Rv2565 | conserved hypothetical protein |

|
 -0.30057 -0.30057
| |
| Rv0285 | PE family protein |

|
 -0.30015 -0.30015
| |
| Rv1597 | hypothetical protein |


|
 -0.30002 -0.30002
|
Genes with positive correlation 
Categories Enriched  : L
: L
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv2601 | spermidine synthase speE |

|
 0.52403 0.52403
| |
| Rv0698 | conserved hypothetical protein |

|
 0.51895 0.51895
| |
| Rv0363c | fructose-bisphosphate aldolase fba |

|
 0.49942 0.49942
| |
| Rv3829c | dehydrogenase |

|
 0.47141 0.47141
| |
| Rv1897c | conserved hypothetical protein |

|
 0.46681 0.46681
| |
| Rv2600 | conserved membrane protein |

|
 0.43265 0.43265
| |
| Rv2670c | conserved hypothetical protein |

|
 0.42635 0.42635
| |
| Rv2716 | conserved hypothetical protein |

|
 0.42566 0.42566
| |
| Rv2807 | conserved hypothetical protein |

|
 0.42113 0.42113
| |
| Rv1170 | N-acetyl-1-D-myo-inosityl-2-amino-2-deoxy-alpha-D-glucopyranoside deacetylase mshB |

|
 0.42101 0.42101
| |
| Rv2089c | dipeptidase pepE |

|
 0.4145 0.4145
| |
| Rv0751c | 3-hydroxyisobutyrate dehydrogenase mmsB |

|
 0.40693 0.40693
| |
| Rv1140 | membrane protein |

|
 0.40476 0.40476
| |
| Rv2014 | transposase |

|
 0.40022 0.40022
| |
| Rv2284 | esterase lipM |

|
 0.39712 0.39712
| |
| Rv3082c | araC/xylS-family virulence-regulating transcriptional regulator virS |

|
 0.39594 0.39594
| |
| Rv3820c | polyketide synthase associated protein papA2 |

|
 0.38843 0.38843
| |
| Rv3347c | PPE family protein |

|
 0.38428 0.38428
| |
| Rv0924c | divalent cation-transport membrane protein mntH |

|
 0.37741 0.37741
| |
| Rv2515c | conserved hypothetical protein |

|
 0.3765 0.3765
| |
| Rv1975 | conserved hypothetical protein |

|
 0.37595 0.37595
| |
| Rv2316 | sugar-transport membrane protein ABC transporter uspA |

|
 0.37505 0.37505
| |
| Rv0797 | transposase |

|
 0.37332 0.37332
| |
| Rv0575c | oxidoreductase |


|
 0.37245 0.37245
| |
| Rv1204c | conserved hypothetical protein |

|
 0.37223 0.37223
| |
| Rv1707 | conserved membrane protein |

|
 0.36755 0.36755
| |
| Rv0806c | UDP-glucose-4-epimerase cpsY |

|
 0.36564 0.36564
| |
| Rv3110 | pterin-4-alpha-carbinolamine dehydratase moaB1 |

|
 0.36396 0.36396
| |
| Rv3185 | transposase |

|
 0.36347 0.36347
| |
| Rv0837c | hypothetical protein |

|
 0.36258 0.36258
| |
| Rv0204c | conserved membrane protein |

|
 0.36108 0.36108
| |
| Rv2985 | hydrolase mutT1 |


|
 0.36049 0.36049
| |
| Rv1499 | hypothetical protein |

|
 0.3604 0.3604
| |
| Rv2026c | conserved hypothetical protein |

|
 0.35967 0.35967
| |
| Rv2290 | lipoprotein lppO |

|
 0.35894 0.35894
| |
| Rv1572c | conserved hypothetical protein |

|
 0.35464 0.35464
| |
| Rv2692 | trk system potassium uptake protein ceoC |

|
 0.35315 0.35315
| |
| Rv0255c | cobyric acid synthase cobQ1 |

|
 0.35176 0.35176
| |
| Rv2644c | hypothetical protein |

|
 0.35133 0.35133
| |
| Rv1716 | conserved hypothetical protein |

|
 0.35003 0.35003
| |
| Rv1675c | transcriptional regulator |

|
 0.34803 0.34803
| |
| Rv0371c | conserved hypothetical protein |

|
 0.34647 0.34647
| |
| Rv3205c | conserved hypothetical protein |

|
 0.34614 0.34614
| |
| Rv0160c | PE family protein |

|
 0.34581 0.34581
| |
| Rv1587c | 13E12 repeat family protein |

|
 0.34526 0.34526
| |
| Rv0728c | D-3-phosphoglycerate dehydrogenase serA2 |

|
 0.34512 0.34512
| |
| Rv2355 | transposase |

|
 0.34484 0.34484
| |
| Rv2066 | bifunctional cobI-cobJ fusion protein |

|
 0.34418 0.34418
| |
| Rv2814c | transposase |

|
 0.34399 0.34399
| |
| Rv0137c | peptide methionine sulfoxide reductase msrA |

|
 0.34396 0.34396
| |
| Rv3775 | lipase lipE |

|
 0.34339 0.34339
| |
| Rv0150c | conserved hypothetical protein |

|
 0.34328 0.34328
| |
| Rv0414c | thiamine-phosphate pyrophosphorylase thiE |

|
 0.34266 0.34266
| |
| Rv3018c | PPE family protein |

|
 0.34171 0.34171
| |
| Rv3635 | conserved membrane protein |

|
 0.33948 0.33948
| |
| Rv1186c | conserved hypothetical protein |


|
 0.33946 0.33946
| |
| Rv0771 | 4-carboxymuconolactone decarboxylase cmd |

|
 0.33848 0.33848
| |
| Rv0592 | MCE-family protein mce2D |

|
 0.33729 0.33729
| |
| Rv2167c | transposase |

|
 0.33571 0.33571
| |
| Rv2479c | transposase |

|
 0.33558 0.33558
| |
| Rv1742 | hypothetical protein |

|
 0.33556 0.33556
| |
| Rv1489c |

|
 0.33533 0.33533
| ||
| Rv0796 | transposase |

|
 0.33499 0.33499
| |
| Rv1777 | cytochrome P450 144 cyp144 |

|
 0.33382 0.33382
| |
| Rv2408 | PE family protein |

|
 0.32974 0.32974
| |
| Rv0355c | PPE family protein |

|
 0.32968 0.32968
| |
| Rv1873 | conserved hypothetical protein |

|
 0.32924 0.32924
| |
| Rv2531c | amino acid decarboxylase |

|
 0.3292 0.3292
| |
| Rv2599 | conserved membrane protein |

|
 0.3288 0.3288
| |
| Rv1759c | PE-PGRS family protein |

|
 0.32788 0.32788
| |
| Rv3112 | molybdenum cofactor biosynthesis protein D moaD1 |

|
 0.32738 0.32738
| |
| Rv1043c | conserved hypothetical protein |

|
 0.32679 0.32679
| |
| Rv0601c | two component system sensor kinase |

|
 0.32595 0.32595
| |
| Rv1039c | PPE family protein |

|
 0.32539 0.32539
| |
| Rv0132c | F420-dependent glucose-6-phosphate dehydrogenase fgd2 |

|
 0.32391 0.32391
| |
| Rv1085c | hemolysin |

|
 0.32287 0.32287
| |
| Rv1290c | conserved hypothetical protein |

|
 0.3227 0.3227
| |
| Rv1020 | transcription-repair coupling factor mfd |


|
 0.32222 0.32222
| |
| Rv0304c | PPE family protein |

|
 0.32164 0.32164
| |
| Rv0498 | conserved hypothetical protein |

|
 0.32128 0.32128
| |
| Rv0344c | lipoprotein lpqJ |

|
 0.32109 0.32109
| |
| Rv1141c | enoyl-CoA hydratase echA11 |

|
 0.32108 0.32108
| |
| Rv3178 | conserved hypothetical protein |

|
 0.32051 0.32051
| |
| Rv2820c | hypothetical protein |

|
 0.32048 0.32048
| |
| Rv0232 | transcriptional regulator, tetR/acrR-family |

|
 0.32045 0.32045
| |
| Rv3333c | hypothetical proline rich protein |

|
 0.32045 0.32045
| |
| Rv1850 | urease alpha subunit ureC |

|
 0.31844 0.31844
| |
| Rv3108 | hypothetical protein |

|
 0.31787 0.31787
| |
| Rv0028 | hypothetical protein |

|
 0.31702 0.31702
| |
| Rv2529 | hypothetical protein |

|
 0.31652 0.31652
| |
| Rv0008c | membrane protein |

|
 0.31642 0.31642
| |
| Rv1949c | conserved hypothetical protein |

|
 0.31545 0.31545
| |
| Rv3912 | hypothetical alanine rich protein |

|
 0.31282 0.31282
| |
| Rv0032 | 8-amino-7-oxononanoate synthase bioF2 |

|
 0.31175 0.31175
| |
| Rv2279 | transposase |

|
 0.31166 0.31166
| |
| Rv0347 | conserved membrane protein |

|
 0.3108 0.3108
| |
| Rv0152c | PE family protein |

|
 0.31026 0.31026
| |
| Rv3845 | hypothetical protein |

|
 0.30977 0.30977
| |
| Rv0098 | conserved hypothetical protein |

|
 0.30973 0.30973
| |
| Rv3770c | hypothetical leucine rich protein |

|
 0.30855 0.30855
| |
| Rv0943c | monooxygenase |

|
 0.30808 0.30808
| |
| Rv3651 | conserved hypothetical protein |

|
 0.30774 0.30774
| |
| Rv0628c | conserved hypothetical protein |

|
 0.30744 0.30744
| |
| Rv0407 | F420-dependent glucose-6-phosphate dehydrogenase fgd1 |

|
 0.30728 0.30728
| |
| Rv1947 | hypothetical protein |

|
 0.30632 0.30632
| |
| Rv2649 | transposase |

|
 0.30574 0.30574
| |
| Rv0094c | conserved hypothetical protein |

|
 0.30479 0.30479
| |
| Rv1538c | L-aparaginase ansA |


|
 0.30453 0.30453
| |
| Rv0828c | deaminase |


|
 0.30423 0.30423
| |
| Rv0361 | conserved membrane protein |

|
 0.30405 0.30405
| |
| Rv0027 | hypothetical protein |

|
 0.30317 0.30317
| |
| Rv0618 | galactose-1-phosphate uridylyltransferase galTa |

|
 0.30288 0.30288
| |
| Rv1961 | hypothetical protein |

|
 0.30274 0.30274
| |
| Rv1192 | hypothetical protein |

|
 0.3014 0.3014
| |
| Rv0403c | membrane protein mmpS1 |

|
 0.30112 0.30112
| |
| Rv1286 | bifunctional sulfate adenyltransferase/adenylylsulfate kinase cysN/cysC |

|
 0.30081 0.30081
| |
| Rv3532 | PPE family protein |

|
 0.30081 0.30081
| |
| Rv0259c | conserved hypothetical protein |

|
 0.30072 0.30072
| |
| Rv0193c | hypothetical protein |

|
 0.30041 0.30041
| |
| Rv1179c | hypothetical protein |

|
 0.30004 0.30004
|