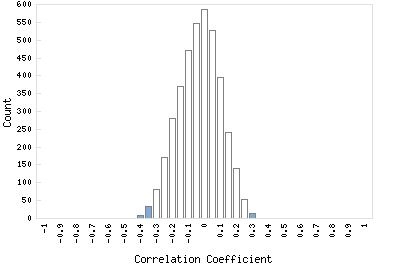
Correlation Catalog M. tuberculosis H37Rv: Rv2637 - transmembrane protein dedA
Distribution of gene correlation: Rv2637 vs. all genes
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv2839c | translation initiation factor IF-2 infB |

|
 -0.40081 -0.40081
| |
| Rv0477 | conserved secreted protein |

|
 -0.3873 -0.3873
| |
| Rv3573c | acyl-CoA dehydrogenase fadE34 |

|
 -0.38007 -0.38007
| |
| Rv2838c | ribosome-binding factor A rbfA |

|
 -0.3744 -0.3744
| |
| Rv1628c | conserved hypothetical protein |

|
 -0.37083 -0.37083
| |
| Rv3183 | transcriptional regulator |

|
 -0.36601 -0.36601
| |
| Rv3504 | acyl-CoA dehydrogenase fadE26 |

|
 -0.3556 -0.3556
| |
| Rv1808 | PPE family protein |

|
 -0.35208 -0.35208
| |
| Rv0197 | oxidoreductase |

|
 -0.35012 -0.35012
| |
| Rv0578c | PE-PGRS family protein |

|
 -0.34947 -0.34947
| |
| Rv2651c | phiRv2 phage protease |

|
 -0.34876 -0.34876
| |
| Rv0476 | conserved membrane protein |

|
 -0.34401 -0.34401
| |
| Rv2650c | phiRv2 phage protein |

|
 -0.34377 -0.34377
| |
| Rv0612 | conserved hypothetical protein |

|
 -0.3432 -0.3432
| |
| Rv1929c | conserved hypothetical protein |

|
 -0.33994 -0.33994
| |
| Rv1807 | PPE family protein |

|
 -0.33887 -0.33887
| |
| Rv3182 | conserved hypothetical protein |

|
 -0.33612 -0.33612
| |
| Rv3537 | dehydrogenase |

|
 -0.33451 -0.33451
| |
| Rv2827c | hypothetical protein |

|
 -0.33002 -0.33002
| |
| Rv3123 | hypothetical protein |

|
 -0.32624 -0.32624
| |
| Rv3917c | chromosome partitioning protein parB |

|
 -0.32424 -0.32424
| |
| Rv3535c | acetaldehyde dehydrogenase |

|
 -0.3189 -0.3189
| |
| Rv0047c | conserved hypothetical protein |

|
 -0.31611 -0.31611
| |
| Rv0841c |

|
 -0.31465 -0.31465
| ||
| Rv2655c | phiRv2 phage protein |

|
 -0.31378 -0.31378
| |
| Rv0244c | acyl-CoA dehydrogenase fadE5 |

|
 -0.31306 -0.31306
| |
| Rv3385c | conserved hypothetical protein |

|
 -0.31193 -0.31193
| |
| Rv3061c | acyl-CoA dehydrogenase fadE22 |

|
 -0.31188 -0.31188
| |
| Rv1420 | excinuclease ABC subunit C uvrC |

|
 -0.31161 -0.31161
| |
| Rv0946c | glucose-6-phosphate isomerase pgi |

|
 -0.31064 -0.31064
| |
| Rv3546 | acetyl-CoA acetyltransferase fadA5 |

|
 -0.3093 -0.3093
| |
| Rv3745c | conserved hypothetical protein |

|
 -0.30865 -0.30865
| |
| Rv2104c | conserved hypothetical protein |

|
 -0.30805 -0.30805
| |
| Rv1809 | PPE family protein |

|
 -0.30733 -0.30733
| |
| Rv2009 | conserved hypothetical protein |

|
 -0.30574 -0.30574
| |
| Rv0123 | hypothetical protein |

|
 -0.30552 -0.30552
| |
| Rv1576c | phiRv1 phage protein |

|
 -0.30455 -0.30455
| |
| Rv0932c | periplasmic phosphate-binding lipoprotein pstS2 |

|
 -0.30419 -0.30419
| |
| Rv2595 | conserved hypothetical protein |

|
 -0.30398 -0.30398
| |
| Rv3189 | conserved hypothetical protein |

|
 -0.30297 -0.30297
| |
| Rv3406 | dioxygenase |

|
 -0.30283 -0.30283
| |
| Rv0196 | transcriptional regulator |

|
 -0.3021 -0.3021
| |
| Rv3595c | PE-PGRS family protein |

|
 -0.30176 -0.30176
| |
| Rv2933 | phenolpthiocerol synthesis type-I polyketide synthase ppsC |

|
 -0.30057 -0.30057
|
Genes with positive correlation 
Categories Enriched  : None
: None
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv1271c | conserved secreted protein |

|
 0.36667 0.36667
| |
| Rv2174 | conserved membrane protein |

|
 0.34728 0.34728
| |
| Rv3332 | N-acetylglucosamine-6-phosphate deacetylase nagA |

|
 0.34358 0.34358
| |
| Rv3690 | conserved membrane protein |

|
 0.34159 0.34159
| |
| Rv3762c | hydrolase |

|
 0.33811 0.33811
| |
| Rv2536 | conserved membrane protein |

|
 0.33632 0.33632
| |
| Rv3734c | conserved hypothetical protein |

|
 0.33381 0.33381
| |
| Rv3691 | conserved hypothetical protein |

|
 0.31586 0.31586
| |
| Rv2683 | conserved hypothetical protein |

|
 0.31408 0.31408
| |
| Rv3718c | conserved hypothetical protein |

|
 0.31406 0.31406
| |
| Rv2409c | conserved hypothetical protein |

|
 0.30626 0.30626
| |
| Rv0118c | oxalyl-CoA decarboxylase oxcA |
![COG0028: (EH) Thiamine pyrophosphate-requiring enzymes [acetolactate synthase; pyruvate dehydrogenase (cytochrome); glyoxylate carboligase; phosphonopyruvate decarboxylase]](supportFiles/E.png)
![COG0028: (EH) Thiamine pyrophosphate-requiring enzymes [acetolactate synthase; pyruvate dehydrogenase (cytochrome); glyoxylate carboligase; phosphonopyruvate decarboxylase]](supportFiles/H.png)
|
 0.30612 0.30612
| |
| Rv1359 | transcriptional regulator |

|
 0.30367 0.30367
| |
| Rv2740 | conserved hypothetical protein |

|
 0.30321 0.30321
| |
| Rv2128 | conserved membrane protein |

|
 0.30181 0.30181
| |
| Rv2343c | DNA primase dnaG |

|
 0.30022 0.30022
|