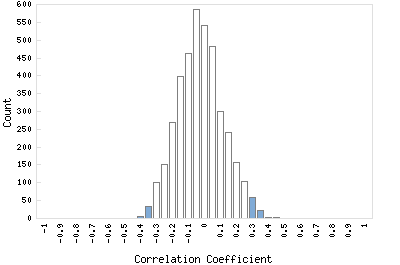
Correlation Catalog M. tuberculosis H37Rv: Rv2085 - conserved hypothetical protein
Distribution of gene correlation: Rv2085 vs. all genes
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv1532c | conserved hypothetical protein |

|
 -0.42102 -0.42102
| |
| Rv2640c | transcriptional regulator, arsR-family |

|
 -0.38739 -0.38739
| |
| Rv2249c | glycerol-3-phosphate dehydrogenase glpD1 |

|
 -0.37002 -0.37002
| |
| Rv3526 | oxidoreductase |


|
 -0.36273 -0.36273
| |
| Rv3504 | acyl-CoA dehydrogenase fadE26 |

|
 -0.3604 -0.3604
| |
| Rv0244c | acyl-CoA dehydrogenase fadE5 |

|
 -0.35343 -0.35343
| |
| Rv0574c | conserved hypothetical protein |

|
 -0.35288 -0.35288
| |
| Rv0211 | iron-regulated phosphoenolpyruvate carboxykinase pckA |

|
 -0.35006 -0.35006
| |
| Rv3574 | transcriptional regulator, tetR-family |

|
 -0.34963 -0.34963
| |
| Rv0467 | isocitrate lyase icl |

|
 -0.34737 -0.34737
| |
| Rv1039c | PPE family protein |

|
 -0.34735 -0.34735
| |
| Rv0148 | short-chain type dehydrogenase/reductase |


|
 -0.34311 -0.34311
| |
| Rv0330c | hypothetical protein |

|
 -0.34269 -0.34269
| |
| Rv0027 | hypothetical protein |

|
 -0.33924 -0.33924
| |
| Rv3515c | fatty-acid-CoA ligase fadD19 |


|
 -0.33582 -0.33582
| |
| Rv1939 | oxidoreductase |

|
 -0.33193 -0.33193
| |
| Rv3337 | conserved hypothetical protein |

|
 -0.33165 -0.33165
| |
| Rv2777c | conserved hypothetical protein |

|
 -0.33109 -0.33109
| |
| Rv3503c | ferredoxin fdxD |

|
 -0.32484 -0.32484
| |
| Rv3538 | dehydrogenase |

|
 -0.32468 -0.32468
| |
| Rv2624c | conserved hypothetical protein |

|
 -0.32406 -0.32406
| |
| Rv0415 | thiamine biosynthesis oxidoreductase thiO |

|
 -0.3237 -0.3237
| |
| Rv1628c | conserved hypothetical protein |

|
 -0.32358 -0.32358
| |
| Rv0147 | aldehyde dehydrogenase NAD-dependent |

|
 -0.32075 -0.32075
| |
| Rv3638 | transposase |

|
 -0.31989 -0.31989
| |
| Rv3338 | conserved hypothetical protein |

|
 -0.31912 -0.31912
| |
| Rv0885 | conserved hypothetical protein |

|
 -0.31875 -0.31875
| |
| Rv0125 | serine protease pepA |

|
 -0.31858 -0.31858
| |
| Rv1637c | conserved hypothetical protein |

|
 -0.31529 -0.31529
| |
| Rv1731 | succinate-semialdehyde dehydrogenase [NADP+]-dependent gabD2 |

|
 -0.31499 -0.31499
| |
| Rv0080 | conserved hypothetical protein |

|
 -0.3147 -0.3147
| |
| Rv0476 | conserved membrane protein |

|
 -0.31419 -0.31419
| |
| Rv0654 | dioxygenase |

|
 -0.30976 -0.30976
| |
| Rv2330c | lipoprotein lppP |

|
 -0.30807 -0.30807
| |
| Rv1161 | respiratory nitrate reductase alpha chain narG |

|
 -0.30722 -0.30722
| |
| Rv2043c | pyrazinamidase/nicotinamidas pncA |

|
 -0.30688 -0.30688
| |
| Rv0489 | phosphoglycerate mutase 1 gpm1 |

|
 -0.30594 -0.30594
| |
| Rv3546 | acetyl-CoA acetyltransferase fadA5 |

|
 -0.30396 -0.30396
| |
| Rv3527 | hypothetical protein |

|
 -0.30296 -0.30296
| |
| Rv3537 | dehydrogenase |

|
 -0.30251 -0.30251
| |
| Rv0475 | iron-regulated heparin binding hemagglutinin hbhA |

|
 -0.30047 -0.30047
|
Genes with positive correlation 
Categories Enriched  : None
: None
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv3158 | NADH dehydrogenase I chain N nuoN |

|
 0.45748 0.45748
| |
| Rv0542c | O-succinylbenzoic acid-CoA ligase menE |


|
 0.45261 0.45261
| |
| Rv2763c | dihydrofolate reductase dfrA |

|
 0.45018 0.45018
| |
| Rv1107c | exodeoxyribonuclease VII small subunit xseB |

|
 0.43964 0.43964
| |
| Rv3148 | NADH dehydrogenase I chain D nuoD |

|
 0.41666 0.41666
| |
| Rv0425c | metal cation transporting P-type ATPase ctpH |

|
 0.41296 0.41296
| |
| Rv3282 | conserved hypothetical protein |

|
 0.40494 0.40494
| |
| Rv3156 | NADH dehydrogenase I chain L nuoL |


|
 0.39905 0.39905
| |
| Rv1016c | lipoprotein lpqT |

|
 0.39684 0.39684
| |
| Rv3680 | anion transporter ATPase |

|
 0.38766 0.38766
| |
| Rv1906c | conserved hypothetical protein |

|
 0.38709 0.38709
| |
| Rv2901c | conserved hypothetical protein |

|
 0.3858 0.3858
| |
| Rv3150 | NADH dehydrogenase I chain F nuoF |

|
 0.38507 0.38507
| |
| Rv2881c | membrane phosphatidate cytidylyltransferase cdsA |

|
 0.38454 0.38454
| |
| Rv3494c | MCE-family protein mce4F |

|
 0.38389 0.38389
| |
| Rv1614 | prolipoprotein diacylglyceryl transferases lgt |

|
 0.37783 0.37783
| |
| Rv3154 | NADH dehydrogenase I chain J nuoJ |

|
 0.37529 0.37529
| |
| Rv2532c | hypothetical protein |

|
 0.37314 0.37314
| |
| Rv3151 | NADH dehydrogenase I chain G nuoG |

|
 0.372 0.372
| |
| Rv0168 | hypothetical membrane protein yrbE1B |

|
 0.36728 0.36728
| |
| Rv3700c | conserved hypothetical protein |

|
 0.36529 0.36529
| |
| Rv1502 | hypothetical protein |

|
 0.36488 0.36488
| |
| Rv1632c | hypothetical protein |

|
 0.362 0.362
| |
| Rv2966c | methyltransferase/methylase |

|
 0.36185 0.36185
| |
| Rv0499 | conserved hypothetical protein |

|
 0.3609 0.3609
| |
| Rv3149 | NADH dehydrogenase I chain E nuoE |

|
 0.35979 0.35979
| |
| Rv2139 | dihydroorotate dehydrogenase pyrD |

|
 0.35806 0.35806
| |
| Rv1547 | DNA polymerase III alpha chain dnaE1 |

|
 0.35187 0.35187
| |
| Rv3920c | hypothetical protein |

|
 0.35149 0.35149
| |
| Rv0111 | transmembrane acyltransferase |

|
 0.34991 0.34991
| |
| Rv1426c | esterase lipO |

|
 0.3484 0.3484
| |
| Rv2755c | Type I restriction/modification system specificity determinant hsdS_1 |

|
 0.34755 0.34755
| |
| Rv0176 | MCE-associated transmembrane protein |

|
 0.34543 0.34543
| |
| Rv1612 | tryptophan synthase, beta subunit trpB |

|
 0.34432 0.34432
| |
| Rv3146 | NADH dehydrogenase I chain B nuoB |

|
 0.34303 0.34303
| |
| Rv1723 | hydrolase |

|
 0.34201 0.34201
| |
| Rv2681 | conserved alanine rich protein |

|
 0.34043 0.34043
| |
| Rv0402c | transmembrane transport protein mmpL1 |

|
 0.33909 0.33909
| |
| Rv0173 | MCE-family lipoprotein mce1E |

|
 0.33741 0.33741
| |
| Rv0541c | conserved membrane protein |

|
 0.33659 0.33659
| |
| Rv3308 | phosphomannomutase pmmB |

|
 0.33528 0.33528
| |
| Rv0166 | fatty-acid-CoA ligase fadD5 |


|
 0.33514 0.33514
| |
| Rv1217c | tetronasin-transport membrane protein ABC transporter |

|
 0.33407 0.33407
| |
| Rv3498c | MCE-family protein mce4B |

|
 0.33393 0.33393
| |
| Rv1613 | tryptophan synthase, alpha subunit trpA |

|
 0.33271 0.33271
| |
| Rv3322c | methyltransferase |


|
 0.33205 0.33205
| |
| Rv1312 | conserved secreted protein |

|
 0.33148 0.33148
| |
| Rv0734 | methionine aminopeptidase mapA |

|
 0.33056 0.33056
| |
| Rv2989 | transcriptional regulator |

|
 0.32922 0.32922
| |
| Rv0170 | MCE-family protein mce1B |

|
 0.32816 0.32816
| |
| Rv1679 | acyl-CoA dehydrogenase fadE16 |

|
 0.32679 0.32679
| |
| Rv2757c | conserved hypothetical protein |

|
 0.32642 0.32642
| |
| Rv1747 | conserved transmembrane ATP-binding protein ABC transporter |

|
 0.32612 0.32612
| |
| Rv0171 | MCE-family protein mce1C |

|
 0.32458 0.32458
| |
| Rv1835c | conserved hypothetical protein |

|
 0.32344 0.32344
| |
| Rv1026 | conserved hypothetical protein |


|
 0.32176 0.32176
| |
| Rv2758c | conserved hypothetical protein |

|
 0.32142 0.32142
| |
| Rv2481c | hypothetical protein |

|
 0.31983 0.31983
| |
| Rv2761c | Type I restriction/modification system specificity determinant hsdS |

|
 0.31909 0.31909
| |
| Rv1618 | acyl-CoA thioesterase II tesB1 |

|
 0.31895 0.31895
| |
| Rv2092c | ATP-dependent DNA helicase helY |



|
 0.31871 0.31871
| |
| Rv0679c | conserved threonine rich protein |

|
 0.31773 0.31773
| |
| Rv2988c | 3-isopropylmalate dehydratase large subunit leuC |

|
 0.31688 0.31688
| |
| Rv3781 | O-antigen/lipopolysaccharide transport ATP-binding protein ABC transporter rfbE |

|
 0.31676 0.31676
| |
| Rv0172 | MCE-family protein mce1D |

|
 0.31638 0.31638
| |
| Rv3496c | MCE-family protein mce4D |

|
 0.31594 0.31594
| |
| Rv0556 | conserved membrane protein |

|
 0.31522 0.31522
| |
| Rv1271c | conserved secreted protein |

|
 0.31511 0.31511
| |
| Rv0783c | multidrug resistance membrane efflux protein emrB |




|
 0.31425 0.31425
| |
| Rv2510c | conserved hypothetical protein |

|
 0.31386 0.31386
| |
| Rv0059 | hypothetical protein |

|
 0.31277 0.31277
| |
| Rv3921c | conserved membrane protein |

|
 0.31276 0.31276
| |
| Rv3321c | conserved hypothetical protein |

|
 0.31229 0.31229
| |
| Rv2083 | conserved hypothetical protein |

|
 0.31143 0.31143
| |
| Rv0225 | conserved hypothetical protein |

|
 0.31059 0.31059
| |
| Rv0200 | conserved membrane protein |

|
 0.31006 0.31006
| |
| Rv0433 | conserved hypothetical protein |

|
 0.30926 0.30926
| |
| Rv1427c | long-chain fatty-acid-CoA ligase fadD12 |


|
 0.30876 0.30876
| |
| Rv0178 | MCE-associated membrane protein |

|
 0.30838 0.30838
| |
| Rv0907 | conserved hypothetical protein |

|
 0.30793 0.30793
| |
| Rv1550 | fatty-acid-CoA ligase fadD11 |

|
 0.30699 0.30699
| |
| Rv1166 | lipoprotein lpqW |


|
 0.30651 0.30651
| |
| Rv0070c | serine hydroxymethyltransferase 2 glyA2 |

|
 0.30651 0.30651
| |
| Rv0050 | bifunctional penicillin-binding protein ponA1 |

|
 0.30637 0.30637
| |
| Rv3152 | NADH dehydrogenase I chain H nuoH |

|
 0.30534 0.30534
| |
| Rv1798 | conserved hypothetical protein |

|
 0.30335 0.30335
| |
| Rv2112c | conserved hypothetical protein |

|
 0.30288 0.30288
| |
| Rv1002c | conserved membrane protein |

|
 0.30149 0.30149
|