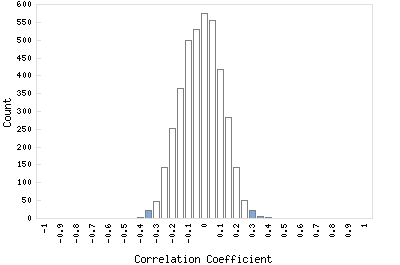
Correlation Catalog M. tuberculosis H37Rv: Rv1914c - hypothetical protein
Distribution of gene correlation: Rv1914c vs. all genes
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv2308 | conserved hypothetical protein |

|
 -0.38959 -0.38959
| |
| Rv2059 | conserved hypothetical protein |

|
 -0.35128 -0.35128
| |
| Rv1570 | dethiobiotin synthetase bioD |

|
 -0.34687 -0.34687
| |
| Rv0074 | conserved hypothetical protein |

|
 -0.34321 -0.34321
| |
| Rv0224c | methyltransferase/methylase |


|
 -0.34131 -0.34131
| |
| Rv0541c | conserved membrane protein |

|
 -0.33928 -0.33928
| |
| Rv2403c | lipoprotein lppR |

|
 -0.33844 -0.33844
| |
| Rv3640c | transposase |

|
 -0.33401 -0.33401
| |
| Rv1743 | transmembrane serine/threonine-protein kinase E pknE |

|
 -0.33025 -0.33025
| |
| Rv3109 | molybdenum cofactor biosynthesis protein A moaA1 |

|
 -0.33015 -0.33015
| |
| Rv3637 | transposase |

|
 -0.3301 -0.3301
| |
| Rv3775 | lipase lipE |

|
 -0.32983 -0.32983
| |
| Rv3107c | alkyldihydroxyacetonephosphate synthase agpS |

|
 -0.32953 -0.32953
| |
| Rv2285 | conserved hypothetical protein |

|
 -0.3286 -0.3286
| |
| Rv3111 | molybdenum cofactor biosynthesis protein C moaC1 |

|
 -0.32849 -0.32849
| |
| Rv0191 | conserved membrane protein |

|
 -0.32692 -0.32692
| |
| Rv3684 | lyase |

|
 -0.32439 -0.32439
| |
| Rv2095c | conserved hypothetical protein |

|
 -0.32305 -0.32305
| |
| Rv0798c | 29 kda antigen cfp29 |

|
 -0.32271 -0.32271
| |
| Rv0825c | conserved hypothetical protein |

|
 -0.32033 -0.32033
| |
| Rv0107c | cation-transporter ATPase I ctpI |

|
 -0.31816 -0.31816
| |
| Rv3359 | oxidoreductase |

|
 -0.31429 -0.31429
| |
| Rv2742c | conserved arginine rich protein |

|
 -0.31094 -0.31094
| |
| Rv1607 | ionic transporter membrane protein chaA |

|
 -0.30804 -0.30804
| |
| Rv0096 | PPE family protein |

|
 -0.3025 -0.3025
|
Genes with positive correlation 
Categories Enriched  : None
: None
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv2798c | conserved hypothetical protein |

|
 0.41833 0.41833
| |
| Rv2304c | hypothetical protein |

|
 0.40237 0.40237
| |
| Rv0756c | hypothetical protein |

|
 0.40173 0.40173
| |
| Rv2420c | conserved hypothetical protein |

|
 0.40013 0.40013
| |
| Rv1890c | hypothetical protein |

|
 0.37894 0.37894
| |
| Rv3382c | lytB-related protein lytB1 |


|
 0.37487 0.37487
| |
| Rv3404c | conserved hypothetical protein |

|
 0.37455 0.37455
| |
| Rv1864c | conserved hypothetical protein |

|
 0.36068 0.36068
| |
| Rv2272 | conserved membrane protein |

|
 0.35735 0.35735
| |
| Rv2432c | hypothetical protein |

|
 0.35578 0.35578
| |
| Rv0134 | epoxide hydrolase ephF |

|
 0.35529 0.35529
| |
| Rv3072c | conserved hypothetical protein |

|
 0.3483 0.3483
| |
| Rv3398c | multifunctional geranylgeranyl pyrophosphate synthetase idsA1 |

|
 0.34441 0.34441
| |
| Rv2300c | conserved hypothetical protein |

|
 0.34098 0.34098
| |
| Rv1512 | nucleotide-sugar epimerase epiA |


|
 0.33861 0.33861
| |
| Rv1848 | urease gamma subunit ureA |

|
 0.33326 0.33326
| |
| Rv3214 | phosphoglycerate mutase gpm2 |

|
 0.3299 0.3299
| |
| Rv2996c | D-3-phosphoglycerate dehydrogenase serA1 |

|
 0.32941 0.32941
| |
| Rv1898 | conserved hypothetical protein |

|
 0.32103 0.32103
| |
| Rv1144 | short-chain type dehydrogenase/reductase |


|
 0.32075 0.32075
| |
| Rv3790 | oxidoreductase |

|
 0.31804 0.31804
| |
| Rv2274c | hypothetical protein |

|
 0.31679 0.31679
| |
| Rv1100 | conserved hypothetical protein |

|
 0.31259 0.31259
| |
| Rv1920 | membrane protein |

|
 0.31258 0.31258
| |
| Rv1609 | anthranilate synthase component I trpE |


|
 0.30945 0.30945
| |
| Rv0562 | polyprenyl-diphosphate synthase grcC1 |

|
 0.3086 0.3086
| |
| Rv2634c | PE-PGRS family protein |

|
 0.30747 0.30747
| |
| Rv0744c | transcriptional regulator |

|
 0.30737 0.30737
| |
| Rv1060 | hypothetical protein |

|
 0.30716 0.30716
| |
| Rv1423 | transcriptional regulator whiA |

|
 0.30519 0.30519
| |
| Rv0780 | phosphoribosylaminoimidazole-succinocarboxamide synthase purC |

|
 0.30203 0.30203
| |
| Rv2222c | glutamine synthetase glnA2 |

|
 0.3019 0.3019
| |
| Rv0740 | conserved hypothetical protein |

|
 0.30174 0.30174
|