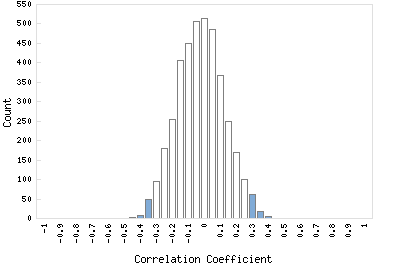
Correlation Catalog M. tuberculosis H37Rv: Rv1443c - hypothetical protein
Distribution of gene correlation: Rv1443c vs. all genes
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0991c | conserved serine rich protein |

|
 -0.41358 -0.41358
| |
| Rv1310 | ATP synthase beta chain atpD |

|
 -0.41262 -0.41262
| |
| Rv1309 | ATP synthase gamma chain atpG |

|
 -0.40575 -0.40575
| |
| Rv2612c | phosphatidylinositol synthase pgsA1 |

|
 -0.39497 -0.39497
| |
| Rv0479c | conserved membrane protein |

|
 -0.38232 -0.38232
| |
| Rv0050 | bifunctional penicillin-binding protein ponA1 |

|
 -0.38136 -0.38136
| |
| Rv1676 | hypothetical protein |

|
 -0.37664 -0.37664
| |
| Rv3014c | NAD-dependent DNA ligase ligA |

|
 -0.36464 -0.36464
| |
| Rv1784 | conserved hypothetical protein |

|
 -0.35324 -0.35324
| |
| Rv2992c | glutamyl-tRNA synthetase gltS |

|
 -0.35165 -0.35165
| |
| Rv2476c | NAD-dependent glutamate dehydrogenase gdh |

|
 -0.34899 -0.34899
| |
| Rv3800c | polyketide synthase pks13 |

|
 -0.34756 -0.34756
| |
| Rv3193c | conserved membrane protein |

|
 -0.34498 -0.34498
| |
| Rv2994 | conserved membrane protein |




|
 -0.34484 -0.34484
| |
| Rv3151 | NADH dehydrogenase I chain G nuoG |

|
 -0.34432 -0.34432
| |
| Rv0143c | conserved membrane protein |

|
 -0.34126 -0.34126
| |
| Rv1916 | isocitrate lyase aceAb |

|
 -0.34028 -0.34028
| |
| Rv0408 | phosphate acetyltransferase pta |

|
 -0.33754 -0.33754
| |
| Rv1796 | proline rich membrane-anchored mycosin mycP5 |

|
 -0.33742 -0.33742
| |
| Rv0934 | periplasmic phosphate-binding lipoprotein pstS1 |

|
 -0.33554 -0.33554
| |
| Rv2967c | pyruvate carboxylase pca |

|
 -0.33421 -0.33421
| |
| Rv3690 | conserved membrane protein |

|
 -0.33301 -0.33301
| |
| Rv0247c | succinate dehydrogenase iron-sulfur subunit |

|
 -0.33241 -0.33241
| |
| Rv1836c | conserved hypothetical protein |

|
 -0.33095 -0.33095
| |
| Rv1604 | inositol-monophosphatase impA |

|
 -0.33007 -0.33007
| |
| Rv1606 | phosphoribosyl-AMP-1,6-cyclohydrolase hisI |

|
 -0.32947 -0.32947
| |
| Rv2112c | conserved hypothetical protein |

|
 -0.32682 -0.32682
| |
| Rv0006 | DNA gyrase subunit A gyrA |

|
 -0.32578 -0.32578
| |
| Rv0058 | replicative DNA helicase dnaB |

|
 -0.3236 -0.3236
| |
| Rv1700 | conserved hypothetical protein |


|
 -0.32148 -0.32148
| |
| Rv3042c | phosphoserine phosphatase serB2 |

|
 -0.31996 -0.31996
| |
| Rv1458c | antibiotic-transport ATP-binding protein ABC transporter |

|
 -0.31962 -0.31962
| |
| Rv2822c | hypothetical protein |

|
 -0.31957 -0.31957
| |
| Rv3051c | ribonucleoside-diphosphate reductase alpha chain nrdE |

|
 -0.31876 -0.31876
| |
| Rv1070c | enoyl-CoA hydratase echA8 |

|
 -0.3184 -0.3184
| |
| Rv0178 | MCE-associated membrane protein |

|
 -0.3158 -0.3158
| |
| Rv2674 | conserved hypothetical protein |

|
 -0.31413 -0.31413
| |
| Rv2610c | alpha-mannosyltransferase pimA |

|
 -0.31327 -0.31327
| |
| Rv2460c | ATP-dependent clp protease proteolytic subunit 2 clpP2 |


|
 -0.31279 -0.31279
| |
| Rv3148 | NADH dehydrogenase I chain D nuoD |

|
 -0.31257 -0.31257
| |
| Rv3030 | conserved hypothetical protein |


|
 -0.31215 -0.31215
| |
| Rv1480 | conserved hypothetical protein |

|
 -0.31132 -0.31132
| |
| Rv1438 | triosephosphate isomerase tpi |

|
 -0.31123 -0.31123
| |
| Rv2614c | threonyl-tRNA synthetase thrS |

|
 -0.31111 -0.31111
| |
| Rv0171 | MCE-family protein mce1C |

|
 -0.31096 -0.31096
| |
| Rv1382 | hypothetical exported protein |

|
 -0.31025 -0.31025
| |
| Rv0503c | cyclopropane-fatty-acyl-phospholipid synthase 2 cmaA2 |

|
 -0.31024 -0.31024
| |
| Rv0346c | L-asparagine permease ansP2 |

|
 -0.30967 -0.30967
| |
| Rv3319 | succinate dehydrogenase iron-sulphur protein subunit sdhB |

|
 -0.30858 -0.30858
| |
| Rv3302c | glycerol-3-phosphate dehydrogenase glpD2 |

|
 -0.30666 -0.30666
| |
| Rv1448c | transaldolase tal |

|
 -0.3059 -0.3059
| |
| Rv2524c | fatty-acid synthase fas |

|
 -0.30543 -0.30543
| |
| Rv3870 | conserved membrane protein |

|
 -0.303 -0.303
| |
| Rv2995c | 3-isopropylmalate dehydrogenase leuB |

|
 -0.30246 -0.30246
| |
| Rv1843c | inosine-5-monophosphate dehydrogenase guaB1 |

|
 -0.30237 -0.30237
| |
| Rv3152 | NADH dehydrogenase I chain H nuoH |

|
 -0.30171 -0.30171
| |
| Rv1304 | ATP synthase A chain atpB |

|
 -0.30061 -0.30061
| |
| Rv2611c | acyltransferase |

|
 -0.30019 -0.30019
| |
| Rv3215 | isochorismate synthase entC |


|
 -0.3 -0.3
|
Genes with positive correlation 
Categories Enriched  : K
: K
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv1044 | conserved hypothetical protein |

|
 0.43534 0.43534
| |
| Rv1267c | transcriptional regulator embR |

|
 0.42695 0.42695
| |
| Rv2827c | hypothetical protein |

|
 0.40744 0.40744
| |
| Rv1960c | conserved hypothetical protein |

|
 0.40124 0.40124
| |
| Rv3183 | transcriptional regulator |

|
 0.39082 0.39082
| |
| Rv1285 | sulfate adenylyltransferase subunit 2 cysD |


|
 0.39056 0.39056
| |
| Rv0275c | transcriptional regulator, tetR-family |

|
 0.39023 0.39023
| |
| Rv3391 | acyl-CoA reductase acrA1 |


|
 0.38202 0.38202
| |
| Rv1081c | conserved membrane protein |

|
 0.37181 0.37181
| |
| Rv1849 | urease beta subunit ureB |

|
 0.36738 0.36738
| |
| Rv1875 | conserved hypothetical protein |

|
 0.36731 0.36731
| |
| Rv2021c | transcriptional regulator |

|
 0.36645 0.36645
| |
| Rv1831 | hypothetical protein |

|
 0.366 0.366
| |
| Rv1806 | PE family protein |

|
 0.35929 0.35929
| |
| Rv0623 | conserved hypothetical protein |

|
 0.35854 0.35854
| |
| Rv0609 | conserved hypothetical protein |

|
 0.35828 0.35828
| |
| Rv0047c | conserved hypothetical protein |

|
 0.35734 0.35734
| |
| Rv3385c | conserved hypothetical protein |

|
 0.35706 0.35706
| |
| Rv2295 | conserved hypothetical protein |

|
 0.35613 0.35613
| |
| Rv2595 | conserved hypothetical protein |

|
 0.35523 0.35523
| |
| Rv0297 | PE-PGRS family protein |

|
 0.35044 0.35044
| |
| Rv1807 | PPE family protein |

|
 0.34906 0.34906
| |
| Rv1951c | conserved hypothetical protein |

|
 0.34595 0.34595
| |
| Rv2517c | hypothetical protein |

|
 0.34337 0.34337
| |
| Rv2643 | arsenic-transport membrane protein arsC |

|
 0.34202 0.34202
| |
| Rv1808 | PPE family protein |

|
 0.34188 0.34188
| |
| Rv0610c | hypothetical protein |

|
 0.34029 0.34029
| |
| Rv2182c | 1-acylglycerol-3-phosphate o-acyltransferase |

|
 0.33692 0.33692
| |
| Rv0137c | peptide methionine sulfoxide reductase msrA |

|
 0.33673 0.33673
| |
| Rv1068c | PE-PGRS family protein |

|
 0.33617 0.33617
| |
| Rv2865 | conserved hypothetical protein |

|
 0.33571 0.33571
| |
| Rv0657c | conserved hypothetical protein |

|
 0.33319 0.33319
| |
| Rv1365c | anti-anti-sigma factor rsfA |

|
 0.33236 0.33236
| |
| Rv0122 | hypothetical protein |

|
 0.33134 0.33134
| |
| Rv2669 | conserved hypothetical protein |


|
 0.33094 0.33094
| |
| Rv3597c | iron-regulated lsr2 protein precursor |

|
 0.33069 0.33069
| |
| Rv1766 | conserved hypothetical protein |

|
 0.3291 0.3291
| |
| Rv2657c | phiRv2 phage protein |

|
 0.32898 0.32898
| |
| Rv2122c | phosphoribosyl-AMP pyrophosphatase hisE |

|
 0.32848 0.32848
| |
| Rv0117 | oxidative stress response regulatory protein oxyS |

|
 0.32681 0.32681
| |
| Rv3917c | chromosome partitioning protein parB |

|
 0.32588 0.32588
| |
| Rv1265 | hypothetical protein |

|
 0.32555 0.32555
| |
| Rv1325c | PE-PGRS family protein |

|
 0.32428 0.32428
| |
| Rv2034 | arsR-type repressor protein |

|
 0.32264 0.32264
| |
| Rv1535 | hypothetical protein |

|
 0.32188 0.32188
| |
| Rv0579 | conserved hypothetical protein |

|
 0.32083 0.32083
| |
| Rv1314c | conserved hypothetical protein |

|
 0.31994 0.31994
| |
| Rv1929c | conserved hypothetical protein |

|
 0.31989 0.31989
| |
| Rv3595c | PE-PGRS family protein |

|
 0.31942 0.31942
| |
| Rv0595c | conserved hypothetical protein |

|
 0.31789 0.31789
| |
| Rv2266 | cytochrome P450 124 cyp124 |

|
 0.31687 0.31687
| |
| Rv1897c | conserved hypothetical protein |

|
 0.31581 0.31581
| |
| Rv2161c | conserved hypothetical protein |

|
 0.31471 0.31471
| |
| Rv0419 | lipoprotein peptidase lpqM |

|
 0.31464 0.31464
| |
| Rv3123 | hypothetical protein |

|
 0.31441 0.31441
| |
| Rv0045c | hydrolase |

|
 0.31376 0.31376
| |
| Rv0369c | membrane oxidoreductase |

|
 0.31363 0.31363
| |
| Rv3180c | hypothetical alanine rich protein |

|
 0.3128 0.3128
| |
| Rv1750c | fatty-acid-CoA ligase fadD1 |


|
 0.31213 0.31213
| |
| Rv2887 | transcriptional regulator |

|
 0.31085 0.31085
| |
| Rv1143 | alpha-methylacyl-CoA racemase mcr |

|
 0.30897 0.30897
| |
| Rv1759c | PE-PGRS family protein |

|
 0.30888 0.30888
| |
| Rv2022c | conserved hypothetical protein |

|
 0.30875 0.30875
| |
| Rv2358 | transcriptional regulator, arsR-family |

|
 0.30847 0.30847
| |
| Rv1990c | transcriptional regulator |

|
 0.3077 0.3077
| |
| Rv1986 | conserved membrane protein |

|
 0.30727 0.30727
| |
| Rv0627 | conserved hypothetical protein |

|
 0.30716 0.30716
| |
| Rv0232 | transcriptional regulator, tetR/acrR-family |

|
 0.30608 0.30608
| |
| Rv3182 | conserved hypothetical protein |

|
 0.30578 0.30578
| |
| Rv2099c | PE family protein |

|
 0.30574 0.30574
| |
| Rv0689c | hypothetical protein |

|
 0.30555 0.30555
| |
| Rv1243c | PE-PGRS family protein |

|
 0.30461 0.30461
| |
| Rv1149 | transposase |

|
 0.30375 0.30375
| |
| Rv2493 | conserved hypothetical protein |

|
 0.30372 0.30372
| |
| Rv2255c | hypothetical protein |

|
 0.30244 0.30244
| |
| Rv1404 | transcriptional regulator |

|
 0.30162 0.30162
| |
| Rv3189 | conserved hypothetical protein |

|
 0.30153 0.30153
| |
| Rv1042c | transposase |

|
 0.30144 0.30144
| |
| Rv1877 | conserved membrane protein |




|
 0.30081 0.30081
| |
| Rv0340 | conserved hypothetical protein |

|
 0.3007 0.3007
| |
| Rv1637c | conserved hypothetical protein |

|
 0.30043 0.30043
| |
| Rv2516c | hypothetical protein |

|
 0.30021 0.30021
| |
| Rv1150 | transposase |

|
 0.3 0.3
|