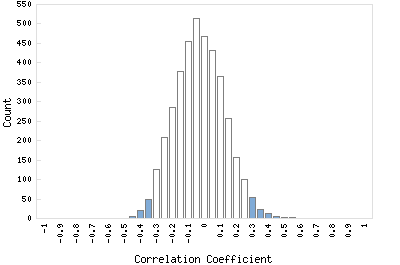
Correlation Catalog M. tuberculosis H37Rv: Rv1065 - conserved hypothetical protein
Distribution of gene correlation: Rv1065 vs. all genes
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv3154 | NADH dehydrogenase I chain J nuoJ |

|
 -0.42422 -0.42422
| |
| Rv3158 | NADH dehydrogenase I chain N nuoN |

|
 -0.42184 -0.42184
| |
| Rv0174 | MCE-family protein mce1F |

|
 -0.41018 -0.41018
| |
| Rv0225 | conserved hypothetical protein |

|
 -0.40675 -0.40675
| |
| Rv3680 | anion transporter ATPase |

|
 -0.39877 -0.39877
| |
| Rv1502 | hypothetical protein |

|
 -0.39689 -0.39689
| |
| Rv1026 | conserved hypothetical protein |


|
 -0.39682 -0.39682
| |
| Rv0170 | MCE-family protein mce1B |

|
 -0.39548 -0.39548
| |
| Rv1632c | hypothetical protein |

|
 -0.39295 -0.39295
| |
| Rv3156 | NADH dehydrogenase I chain L nuoL |


|
 -0.39044 -0.39044
| |
| Rv0910 | conserved hypothetical protein |

|
 -0.38855 -0.38855
| |
| Rv0556 | conserved membrane protein |

|
 -0.38777 -0.38777
| |
| Rv3496c | MCE-family protein mce4D |

|
 -0.37962 -0.37962
| |
| Rv3078 | hydroxylaminobenzene mutase hab |

|
 -0.3793 -0.3793
| |
| Rv0172 | MCE-family protein mce1D |

|
 -0.37594 -0.37594
| |
| Rv0173 | MCE-family lipoprotein mce1E |

|
 -0.37533 -0.37533
| |
| Rv1427c | long-chain fatty-acid-CoA ligase fadD12 |


|
 -0.3623 -0.3623
| |
| Rv0059 | hypothetical protein |

|
 -0.36081 -0.36081
| |
| Rv1230c | membrane protein |

|
 -0.35975 -0.35975
| |
| Rv2074 | conserved hypothetical protein |

|
 -0.35732 -0.35732
| |
| Rv1426c | esterase lipO |

|
 -0.35424 -0.35424
| |
| Rv3498c | MCE-family protein mce4B |

|
 -0.35283 -0.35283
| |
| Rv0542c | O-succinylbenzoic acid-CoA ligase menE |


|
 -0.35278 -0.35278
| |
| Rv3146 | NADH dehydrogenase I chain B nuoB |

|
 -0.35204 -0.35204
| |
| Rv0108c | hypothetical protein |

|
 -0.35135 -0.35135
| |
| Rv2238c | peroxiredoxin ahpE |

|
 -0.34997 -0.34997
| |
| Rv1906c | conserved hypothetical protein |

|
 -0.34661 -0.34661
| |
| Rv0430 | conserved hypothetical protein |

|
 -0.34572 -0.34572
| |
| Rv1547 | DNA polymerase III alpha chain dnaE1 |

|
 -0.34321 -0.34321
| |
| Rv3495c | MCE-family lipoprotein mce4E |

|
 -0.34127 -0.34127
| |
| Rv0783c | multidrug resistance membrane efflux protein emrB |




|
 -0.33984 -0.33984
| |
| Rv0178 | MCE-associated membrane protein |

|
 -0.33835 -0.33835
| |
| Rv3780 | conserved hypothetical protein |

|
 -0.33764 -0.33764
| |
| Rv3155 | NADH dehydrogenase I chain K nuoK |

|
 -0.33752 -0.33752
| |
| Rv0433 | conserved hypothetical protein |

|
 -0.33572 -0.33572
| |
| Rv1614 | prolipoprotein diacylglyceryl transferases lgt |

|
 -0.33484 -0.33484
| |
| Rv3692 | methanol dehydrogenase transcriptional regulator moxR2 |

|
 -0.33437 -0.33437
| |
| Rv2681 | conserved alanine rich protein |

|
 -0.3339 -0.3339
| |
| Rv0499 | conserved hypothetical protein |

|
 -0.33253 -0.33253
| |
| Rv0169 | MCE-family protein mce1A |

|
 -0.33177 -0.33177
| |
| Rv1612 | tryptophan synthase, beta subunit trpB |

|
 -0.33146 -0.33146
| |
| Rv2761c | Type I restriction/modification system specificity determinant hsdS |

|
 -0.33021 -0.33021
| |
| Rv3852 | histone-like protein hns |

|
 -0.3298 -0.3298
| |
| Rv3494c | MCE-family protein mce4F |

|
 -0.32967 -0.32967
| |
| Rv2680 | conserved hypothetical protein |

|
 -0.32961 -0.32961
| |
| Rv3150 | NADH dehydrogenase I chain F nuoF |

|
 -0.32947 -0.32947
| |
| Rv0177 | MCE-associated protein |

|
 -0.32745 -0.32745
| |
| Rv1626 | two component system transcriptional regulator |

|
 -0.32725 -0.32725
| |
| Rv3152 | NADH dehydrogenase I chain H nuoH |

|
 -0.32691 -0.32691
| |
| Rv1446c | oxpp cycle protein opcA |

|
 -0.32548 -0.32548
| |
| Rv1770 | conserved hypothetical protein |

|
 -0.32376 -0.32376
| |
| Rv3782 | L-rhamnosyltransferase |

|
 -0.32363 -0.32363
| |
| Rv3792 | conserved membrane protein |

|
 -0.32321 -0.32321
| |
| Rv3157 | NADH dehydrogenase I chain M nuoM |

|
 -0.32156 -0.32156
| |
| Rv1754c | conserved hypothetical protein |

|
 -0.32038 -0.32038
| |
| Rv0171 | MCE-family protein mce1C |

|
 -0.31904 -0.31904
| |
| Rv3690 | conserved membrane protein |

|
 -0.31877 -0.31877
| |
| Rv3648c | cold shock protein A cspA |

|
 -0.31657 -0.31657
| |
| Rv0409 | acetate kinase ackA |

|
 -0.31652 -0.31652
| |
| Rv2228c | conserved hypothetical protein |

|
 -0.31642 -0.31642
| |
| Rv0444c | conserved hypothetical protein |

|
 -0.31515 -0.31515
| |
| Rv0890c | transcriptional regulator, luxR-family |

|
 -0.31383 -0.31383
| |
| Rv2763c | dihydrofolate reductase dfrA |

|
 -0.31347 -0.31347
| |
| Rv3438 | conserved hypothetical protein |

|
 -0.30984 -0.30984
| |
| Rv3262 | F420 biosynthesis protein fbiB |

|
 -0.30916 -0.30916
| |
| Rv3200c | transmembrane cation transporter |

|
 -0.30837 -0.30837
| |
| Rv1882c | short-chain type dehydrogenase/reductase |


|
 -0.30696 -0.30696
| |
| Rv0346c | L-asparagine permease ansP2 |

|
 -0.30675 -0.30675
| |
| Rv0546c | conserved hypothetical protein |

|
 -0.3058 -0.3058
| |
| Rv1883c | conserved hypothetical protein |

|
 -0.30545 -0.30545
| |
| Rv1920 | membrane protein |

|
 -0.30469 -0.30469
| |
| Rv3477 | PE family protein |

|
 -0.30301 -0.30301
| |
| Rv2840c | conserved hypothetical protein |

|
 -0.30149 -0.30149
| |
| Rv1166 | lipoprotein lpqW |


|
 -0.3013 -0.3013
|
Genes with positive correlation 
Categories Enriched  : P
: P
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv1066 | conserved hypothetical protein |

|
 0.78979 0.78979
| |
| Rv1790 | PPE family protein |

|
 0.59588 0.59588
| |
| Rv0485 | transcriptional regulator |

|
 0.55076 0.55076
| |
| Rv1787 | PPE family protein |

|
 0.54442 0.54442
| |
| Rv3338 | conserved hypothetical protein |

|
 0.52116 0.52116
| |
| Rv2400c | sulfate-binding lipoprotein subI |

|
 0.49028 0.49028
| |
| Rv2030c | conserved hypothetical protein |

|
 0.47206 0.47206
| |
| Rv2624c | conserved hypothetical protein |

|
 0.47105 0.47105
| |
| Rv2025c | conserved membrane protein |

|
 0.46993 0.46993
| |
| Rv1733c | conserved membrane protein |

|
 0.46317 0.46317
| |
| Rv0571c | conserved hypothetical protein |

|
 0.43304 0.43304
| |
| Rv2005c | conserved hypothetical protein |

|
 0.42892 0.42892
| |
| Rv2623 | conserved hypothetical protein |

|
 0.4275 0.4275
| |
| Rv3416 | transcriptional regulator whib-like whiB3 |

|
 0.42499 0.42499
| |
| Rv2667 | ATP-dependent protease ATP-binding subunit clpC2 |

|
 0.4247 0.4247
| |
| Rv3133c | two component system transcriptional regulator devR |


|
 0.42094 0.42094
| |
| Rv2631 | conserved hypothetical protein |

|
 0.41798 0.41798
| |
| Rv2028c | conserved hypothetical protein |

|
 0.41273 0.41273
| |
| Rv2628 | hypothetical protein |

|
 0.41131 0.41131
| |
| Rv1734c | conserved hypothetical protein |

|
 0.41101 0.41101
| |
| Rv2004c | conserved hypothetical protein |

|
 0.40912 0.40912
| |
| Rv1089 | PE family protein |

|
 0.40715 0.40715
| |
| Rv3033 | hypothetical protein |

|
 0.40583 0.40583
| |
| Rv1403c | methyltransferase |


|
 0.40148 0.40148
| |
| Rv2398c | sulfate-transport membrane protein ABC transporter cysW |

|
 0.3994 0.3994
| |
| Rv0390 | conserved hypothetical protein |

|
 0.39865 0.39865
| |
| Rv1736c | nitrate reductase narX |

|
 0.39572 0.39572
| |
| Rv0467 | isocitrate lyase icl |

|
 0.39557 0.39557
| |
| Rv1082 | mycothiol conjugate amidase mca |

|
 0.39243 0.39243
| |
| Rv1284 | conserved hypothetical protein |

|
 0.39223 0.39223
| |
| Rv3289c | transmembrane protein |

|
 0.38895 0.38895
| |
| Rv1992c | metal cation transporting P-type ATPase ctpG |

|
 0.38893 0.38893
| |
| Rv1994c | transcriptional regulator |

|
 0.38835 0.38835
| |
| Rv3134c | conserved hypothetical protein |

|
 0.37827 0.37827
| |
| Rv3009c | glutamyl-tRNA(gln) amidotransferase subunit B gatB |

|
 0.37594 0.37594
| |
| Rv0570 | ribonucleoside-diphosphate reductase large subunit nrdZ |

|
 0.37111 0.37111
| |
| Rv1405c | methyltransferase |


|
 0.36886 0.36886
| |
| Rv1998c | conserved hypothetical protein |

|
 0.36717 0.36717
| |
| Rv2666 | transposase |

|
 0.36579 0.36579
| |
| Rv2995c | 3-isopropylmalate dehydrogenase leuB |

|
 0.36484 0.36484
| |
| Rv2029c | phosphofructokinase pfkB |

|
 0.36051 0.36051
| |
| Rv2629 | conserved hypothetical protein |

|
 0.35907 0.35907
| |
| Rv2399c | sulfate-transport membrane protein ABC transporter cysT |

|
 0.35903 0.35903
| |
| Rv2630 | hypothetical protein |

|
 0.35739 0.35739
| |
| Rv0080 | conserved hypothetical protein |

|
 0.35466 0.35466
| |
| Rv0331 | dehydrogenase/reductase |

|
 0.35409 0.35409
| |
| Rv1737c | nitrate/nitrite transporter narK2 |

|
 0.35164 0.35164
| |
| Rv1810 | conserved hypothetical protein |

|
 0.34673 0.34673
| |
| Rv2640c | transcriptional regulator, arsR-family |

|
 0.34546 0.34546
| |
| Rv0476 | conserved membrane protein |

|
 0.34542 0.34542
| |
| Rv0747 | PE-PGRS family protein |

|
 0.34471 0.34471
| |
| Rv0634c | glyoxalase II |

|
 0.34275 0.34275
| |
| Rv2626c | conserved hypothetical protein |

|
 0.34072 0.34072
| |
| Rv0307c | hypothetical protein |

|
 0.33923 0.33923
| |
| Rv2625c | conserved alanine and leucine rich membrane protein |

|
 0.33825 0.33825
| |
| Rv1419 | hypothetical protein |

|
 0.33651 0.33651
| |
| Rv2627c | conserved hypothetical protein |

|
 0.3352 0.3352
| |
| Rv3766 | hypothetical protein |

|
 0.3342 0.3342
| |
| Rv3007c | oxidoreductase |

|
 0.33364 0.33364
| |
| Rv2043c | pyrazinamidase/nicotinamidas pncA |

|
 0.33263 0.33263
| |
| Rv3129 | conserved hypothetical protein |

|
 0.33259 0.33259
| |
| Rv3117 | thiosulfate sulfurtransferase cysA3 |

|
 0.32777 0.32777
| |
| Rv2863 | conserved hypothetical protein |

|
 0.32712 0.32712
| |
| Rv3124 | transcriptional regulator |

|
 0.32709 0.32709
| |
| Rv2643 | arsenic-transport membrane protein arsC |

|
 0.32451 0.32451
| |
| Rv2032 | hypothetical protein acg |

|
 0.32415 0.32415
| |
| Rv3130c | conserved hypothetical protein |

|
 0.32365 0.32365
| |
| Rv0840c | proline iminopeptidase pip |

|
 0.32346 0.32346
| |
| Rv3023c | transposase |

|
 0.32253 0.32253
| |
| Rv2777c | conserved hypothetical protein |

|
 0.32242 0.32242
| |
| Rv0415 | thiamine biosynthesis oxidoreductase thiO |

|
 0.32135 0.32135
| |
| Rv2991 | conserved hypothetical protein |

|
 0.32109 0.32109
| |
| Rv0814c | hypothetical protein sseC2 |

|
 0.31972 0.31972
| |
| Rv3337 | conserved hypothetical protein |

|
 0.31957 0.31957
| |
| Rv3207c | conserved hypothetical protein |

|
 0.31931 0.31931
| |
| Rv3567c | oxidoreductase |

|
 0.31836 0.31836
| |
| Rv3123 | hypothetical protein |

|
 0.31824 0.31824
| |
| Rv1812c | dehydrogenase |

|
 0.31627 0.31627
| |
| Rv0654 | dioxygenase |

|
 0.31563 0.31563
| |
| Rv3290c | L-lysine-epsilon aminotransferase lat |

|
 0.31506 0.31506
| |
| Rv1735c | hypothetical membrane protein |

|
 0.31476 0.31476
| |
| Rv2172c | conserved hypothetical protein |

|
 0.31442 0.31442
| |
| Rv2359 | ferric uptake regulation protein furB |

|
 0.31428 0.31428
| |
| Rv0065 | conserved hypothetical protein |

|
 0.31409 0.31409
| |
| Rv1956 | transcriptional regulator |

|
 0.31258 0.31258
| |
| Rv2651c | phiRv2 phage protease |

|
 0.31237 0.31237
| |
| Rv1392 | S-adenosylmethionine synthetase metK |

|
 0.31104 0.31104
| |
| Rv3118 | hypothetical protein sseC1 |

|
 0.31026 0.31026
| |
| Rv3115 | transposase |

|
 0.30941 0.30941
| |
| Rv0397 | 13E12 repeat family protein |

|
 0.30593 0.30593
| |
| Rv3538 | dehydrogenase |

|
 0.30573 0.30573
| |
| Rv0574c | conserved hypothetical protein |

|
 0.30523 0.30523
| |
| Rv2397c | sulfate-transport ATP-binding protein ABC transporter cysA1 |

|
 0.30452 0.30452
| |
| Rv2642 | transcriptional regulator, arsR-family |

|
 0.30446 0.30446
| |
| Rv1628c | conserved hypothetical protein |

|
 0.30422 0.30422
| |
| Rv3880c | conserved hypothetical protein |

|
 0.30285 0.30285
| |
| Rv1993c | conserved hypothetical protein |

|
 0.30281 0.30281
| |
| Rv0147 | aldehyde dehydrogenase NAD-dependent |

|
 0.30273 0.30273
| |
| Rv2660c | hypothetical protein |

|
 0.30175 0.30175
| |
| Rv0572c | hypothetical protein |

|
 0.30099 0.30099
| |
| Rv0815c | thiosulfate sulfurtransferase cysA2 |

|
 0.3004 0.3004
|