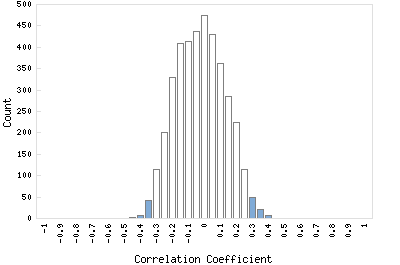
Correlation Catalog M. tuberculosis H37Rv: Rv0776c - conserved hypothetical protein
Distribution of gene correlation: Rv0776c vs. all genes
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv3158 | NADH dehydrogenase I chain N nuoN |

|
 -0.41497 -0.41497
| |
| Rv3208 | transcriptional regulator, tetR-family |

|
 -0.41291 -0.41291
| |
| Rv3272 | conserved hypothetical protein |

|
 -0.39296 -0.39296
| |
| Rv0910 | conserved hypothetical protein |

|
 -0.38734 -0.38734
| |
| Rv2532c | hypothetical protein |

|
 -0.37117 -0.37117
| |
| Rv0542c | O-succinylbenzoic acid-CoA ligase menE |


|
 -0.36237 -0.36237
| |
| Rv2238c | peroxiredoxin ahpE |

|
 -0.36102 -0.36102
| |
| Rv1166 | lipoprotein lpqW |


|
 -0.35335 -0.35335
| |
| Rv3648c | cold shock protein A cspA |

|
 -0.35082 -0.35082
| |
| Rv3151 | NADH dehydrogenase I chain G nuoG |

|
 -0.34967 -0.34967
| |
| Rv1836c | conserved hypothetical protein |

|
 -0.3494 -0.3494
| |
| Rv1770 | conserved hypothetical protein |

|
 -0.34843 -0.34843
| |
| Rv3150 | NADH dehydrogenase I chain F nuoF |

|
 -0.34804 -0.34804
| |
| Rv3154 | NADH dehydrogenase I chain J nuoJ |

|
 -0.34719 -0.34719
| |
| Rv1626 | two component system transcriptional regulator |

|
 -0.34713 -0.34713
| |
| Rv2883c | uridylate kinase pyrH |

|
 -0.34256 -0.34256
| |
| Rv1426c | esterase lipO |

|
 -0.33666 -0.33666
| |
| Rv3149 | NADH dehydrogenase I chain E nuoE |

|
 -0.33601 -0.33601
| |
| Rv3782 | L-rhamnosyltransferase |

|
 -0.33583 -0.33583
| |
| Rv0954 | conserved membrane protein |

|
 -0.33532 -0.33532
| |
| Rv3496c | MCE-family protein mce4D |

|
 -0.33418 -0.33418
| |
| Rv3157 | NADH dehydrogenase I chain M nuoM |

|
 -0.33184 -0.33184
| |
| Rv3680 | anion transporter ATPase |

|
 -0.33067 -0.33067
| |
| Rv3148 | NADH dehydrogenase I chain D nuoD |

|
 -0.32925 -0.32925
| |
| Rv0430 | conserved hypothetical protein |

|
 -0.32704 -0.32704
| |
| Rv3498c | MCE-family protein mce4B |

|
 -0.32549 -0.32549
| |
| Rv3155 | NADH dehydrogenase I chain K nuoK |

|
 -0.3232 -0.3232
| |
| Rv3474 | transposase |

|
 -0.32297 -0.32297
| |
| Rv3186 | transposase |

|
 -0.32021 -0.32021
| |
| Rv0018c | serine/threonine phosphatase ppp |

|
 -0.31894 -0.31894
| |
| Rv1754c | conserved hypothetical protein |

|
 -0.31594 -0.31594
| |
| Rv3116 | molybdenum cofactor biosynthesis protein moeB2 |

|
 -0.31571 -0.31571
| |
| Rv1309 | ATP synthase gamma chain atpG |

|
 -0.31426 -0.31426
| |
| Rv3624c | hypoxanthine-guanine phosphoribosyltransferase hpt |

|
 -0.31327 -0.31327
| |
| Rv0994 | molybdopterin biosynthesis protein moeA1 |

|
 -0.31198 -0.31198
| |
| Rv2152c | UDP-N-acetylmuramate-alanine ligase murC |

|
 -0.31099 -0.31099
| |
| Rv2881c | membrane phosphatidate cytidylyltransferase cdsA |

|
 -0.30811 -0.30811
| |
| Rv3258c | conserved hypothetical protein |

|
 -0.30806 -0.30806
| |
| Rv2418c | hypothetical protein |

|
 -0.30781 -0.30781
| |
| Rv2138 | lipoprotein lppL |

|
 -0.30695 -0.30695
| |
| Rv1502 | hypothetical protein |

|
 -0.30662 -0.30662
| |
| Rv2537c | 3-dehydroquinate dehydratase aroD |

|
 -0.30656 -0.30656
| |
| Rv1436 | glyceraldehyde 3-phosphate dehydrogenase gap |

|
 -0.30458 -0.30458
| |
| Rv3146 | NADH dehydrogenase I chain B nuoB |

|
 -0.30456 -0.30456
| |
| Rv0932c | periplasmic phosphate-binding lipoprotein pstS2 |

|
 -0.30437 -0.30437
| |
| Rv0931c | transmembrane serine/threonine-protein kinase D pknD |

|
 -0.30245 -0.30245
| |
| Rv2074 | conserved hypothetical protein |

|
 -0.30226 -0.30226
| |
| Rv1677 | lipoprotein dsbF |


|
 -0.30202 -0.30202
| |
| Rv0659c | conserved hypothetical protein |

|
 -0.30176 -0.30176
| |
| Rv3028c | electron transfer flavoprotein alpha subunit fixB |

|
 -0.30084 -0.30084
| |
| Rv2271 | conserved hypothetical protein |

|
 -0.30072 -0.30072
|
Genes with positive correlation 
Categories Enriched  : None
: None
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0031 | transposase |

|
 0.46859 0.46859
| |
| Rv0603 | hypothetical exported protein |

|
 0.42952 0.42952
| |
| Rv0817c | hypothetical exported protein |

|
 0.42254 0.42254
| |
| Rv0899 | outer membrane protein A ompA |

|
 0.41754 0.41754
| |
| Rv0388c | PPE family protein |

|
 0.41656 0.41656
| |
| Rv1998c | conserved hypothetical protein |

|
 0.41288 0.41288
| |
| Rv0084 | formate hydrogenlyase hycD |

|
 0.40926 0.40926
| |
| Rv0553 | muconate cycloisomerase menC |

|
 0.40314 0.40314
| |
| Rv0121c | conserved hypothetical protein |

|
 0.40005 0.40005
| |
| Rv0415 | thiamine biosynthesis oxidoreductase thiO |

|
 0.39371 0.39371
| |
| Rv0893c | conserved hypothetical protein |

|
 0.39244 0.39244
| |
| Rv1542c | hemoglobin glbN |

|
 0.38818 0.38818
| |
| Rv2089c | dipeptidase pepE |

|
 0.38667 0.38667
| |
| Rv0392c | membrane NADH dehydrogenase ndhA |

|
 0.38414 0.38414
| |
| Rv2317 | sugar-transport membrane protein ABC transporter uspB |

|
 0.38366 0.38366
| |
| Rv0396 | hypothetical protein |

|
 0.3789 0.3789
| |
| Rv1154c | hypothetical protein |

|
 0.37496 0.37496
| |
| Rv0797 | transposase |

|
 0.37329 0.37329
| |
| Rv0375c | carbon monoxyde dehydrogenase medium chain |

|
 0.36593 0.36593
| |
| Rv1755c | phospholipase C 4 plcD |

|
 0.36551 0.36551
| |
| Rv0915c | PPE family protein |

|
 0.36531 0.36531
| |
| Rv1714 | oxidoreductase |


|
 0.36295 0.36295
| |
| Rv1039c | PPE family protein |

|
 0.36154 0.36154
| |
| Rv0976c | conserved hypothetical protein |

|
 0.35787 0.35787
| |
| Rv0104 | conserved hypothetical protein |

|
 0.35737 0.35737
| |
| Rv1931c | transcriptional regulator |

|
 0.35554 0.35554
| |
| Rv1255c | transcriptional regulator |

|
 0.35328 0.35328
| |
| Rv0395 | hypothetical protein |

|
 0.35261 0.35261
| |
| Rv0267 | membrane nitrite extrusion protein narU |

|
 0.35176 0.35176
| |
| Rv1995 | hypothetical protein |

|
 0.35013 0.35013
| |
| Rv0800 | aminopeptidase pepC |

|
 0.34651 0.34651
| |
| Rv0895 | conserved hypothetical protein |

|
 0.34551 0.34551
| |
| Rv0372c | conserved hypothetical protein |

|
 0.3444 0.3444
| |
| Rv0770 | dehydrogenase/reductase |

|
 0.34366 0.34366
| |
| Rv0521c |

|
 0.34291 0.34291
| ||
| Rv0087 | formate hydrogenase hycE |

|
 0.33933 0.33933
| |
| Rv0099 | fatty-acid-CoA ligase fadD10 |


|
 0.33574 0.33574
| |
| Rv2863 | conserved hypothetical protein |

|
 0.33521 0.33521
| |
| Rv1781c | 4-alpha-glucanotransferase malQ |

|
 0.33509 0.33509
| |
| Rv0943c | monooxygenase |

|
 0.33473 0.33473
| |
| Rv1116 | hypothetical protein |

|
 0.3334 0.3334
| |
| Rv0027 | hypothetical protein |

|
 0.33043 0.33043
| |
| Rv0963c | conserved hypothetical protein |

|
 0.32612 0.32612
| |
| Rv1777 | cytochrome P450 144 cyp144 |

|
 0.32528 0.32528
| |
| Rv2332 | NAD-dependent malate oxidoreductase mez |

|
 0.32494 0.32494
| |
| Rv1089 | PE family protein |

|
 0.32426 0.32426
| |
| Rv1761c | hypothetical exported protein |

|
 0.32016 0.32016
| |
| Rv1228 | lipoprotein lpqX |

|
 0.31803 0.31803
| |
| Rv1972 | MCE-associated membrane protein |

|
 0.31777 0.31777
| |
| Rv3845 | hypothetical protein |

|
 0.31625 0.31625
| |
| Rv1330c | conserved hypothetical protein |

|
 0.31598 0.31598
| |
| Rv0787 | hypothetical protein |

|
 0.31419 0.31419
| |
| Rv2388c | oxygen-independent coproporphyrinogen III oxidase hemN |

|
 0.31408 0.31408
| |
| Rv0052 | conserved hypothetical protein |

|
 0.31298 0.31298
| |
| Rv1204c | conserved hypothetical protein |

|
 0.31291 0.31291
| |
| Rv0420c | transmembrane protein |

|
 0.31245 0.31245
| |
| Rv1969 | MCE-family protein mce3D |

|
 0.312 0.312
| |
| Rv0416 | hypothetical protein thiS |

|
 0.3114 0.3114
| |
| Rv1249c | membrane protein |

|
 0.31096 0.31096
| |
| Rv0465c | transcriptional regulator |

|
 0.31054 0.31054
| |
| Rv0771 | 4-carboxymuconolactone decarboxylase cmd |

|
 0.31049 0.31049
| |
| Rv0150c | conserved hypothetical protein |

|
 0.31041 0.31041
| |
| Rv0812 | amino acid aminotransferase |


|
 0.309 0.309
| |
| Rv0253 | nitrite reductase small subunit nirD |


|
 0.30769 0.30769
| |
| Rv1031 | potassium-transporting ATPase C chain kdpC |

|
 0.30763 0.30763
| |
| Rv0843 | dehydrogenase |

|
 0.30613 0.30613
| |
| Rv0080 | conserved hypothetical protein |

|
 0.30365 0.30365
| |
| Rv1946c | lipoprotein lppG |

|
 0.30284 0.30284
| |
| Rv0675 | enoyl-CoA hydratase echA5 |

|
 0.30262 0.30262
| |
| Rv1203c | hypothetical protein |

|
 0.3025 0.3025
| |
| Rv2623 | conserved hypothetical protein |

|
 0.30198 0.30198
| |
| Rv0034 | conserved hypothetical protein |

|
 0.30164 0.30164
| |
| Rv0449c | conserved hypothetical protein |

|
 0.3013 0.3013
| |
| Rv1733c | conserved membrane protein |

|
 0.30128 0.30128
| |
| Rv3097c | PE-PGRS family protein |

|
 0.30059 0.30059
| |
| Rv0650 | sugar kinase |

|
 0.30027 0.30027
| |
| Rv2679 | enoyl-CoA hydratase echA15 |

|
 0.30024 0.30024
| |
| Rv0101 | peptide synthetase nrp |

|
 0.30007 0.30007
|