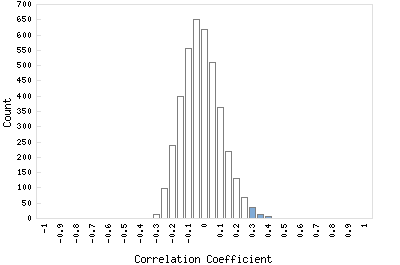
Correlation Catalog M. tuberculosis H37Rv: Rv0507 - transmembrane transport protein mmpL2
Distribution of gene correlation: Rv0507 vs. all genes
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|
Genes with positive correlation 
Categories Enriched  : L
: L
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0032 | 8-amino-7-oxononanoate synthase bioF2 |

|
 0.47758 0.47758
| |
| Rv0152c | PE family protein |

|
 0.44397 0.44397
| |
| Rv0098 | conserved hypothetical protein |

|
 0.43315 0.43315
| |
| Rv0034 | conserved hypothetical protein |

|
 0.42795 0.42795
| |
| Rv0101 | peptide synthetase nrp |

|
 0.42359 0.42359
| |
| Rv0796 | transposase |

|
 0.42062 0.42062
| |
| Rv0396 | hypothetical protein |

|
 0.41917 0.41917
| |
| Rv0394c | secreted protein |

|
 0.4131 0.4131
| |
| Rv0895 | conserved hypothetical protein |

|
 0.39559 0.39559
| |
| Rv0771 | 4-carboxymuconolactone decarboxylase cmd |

|
 0.39051 0.39051
| |
| Rv0395 | hypothetical protein |

|
 0.38583 0.38583
| |
| Rv0151c | PE family protein |

|
 0.38163 0.38163
| |
| Rv0099 | fatty-acid-CoA ligase fadD10 |


|
 0.37966 0.37966
| |
| Rv0121c | conserved hypothetical protein |

|
 0.3742 0.3742
| |
| Rv0193c | hypothetical protein |

|
 0.36979 0.36979
| |
| Rv0414c | thiamine-phosphate pyrophosphorylase thiE |

|
 0.36684 0.36684
| |
| Rv0795 | transposase |

|
 0.36219 0.36219
| |
| Rv0035 | fatty-acid-CoA ligase fadD34 |


|
 0.35676 0.35676
| |
| Rv0012 | conserved membrane protein |

|
 0.35596 0.35596
| |
| Rv0894 | transcriptional regulator, luxR-family |

|
 0.35381 0.35381
| |
| Rv0131c | acyl-CoA dehydrogenase fadE1 |

|
 0.35184 0.35184
| |
| Rv2479c | transposase |

|
 0.35078 0.35078
| |
| Rv0828c | deaminase |


|
 0.3465 0.3465
| |
| Rv1548c | PPE family protein |

|
 0.34644 0.34644
| |
| Rv0797 | transposase |

|
 0.34624 0.34624
| |
| Rv0100 | conserved hypothetical protein |

|
 0.34563 0.34563
| |
| Rv0030 | hypothetical protein |

|
 0.34437 0.34437
| |
| Rv0573c | conserved hypothetical protein |

|
 0.34149 0.34149
| |
| Rv0246 | conserved membrane protein |

|
 0.34089 0.34089
| |
| Rv0105c | 50S ribosomal protein rpmB1 |

|
 0.34022 0.34022
| |
| Rv2545 | conserved hypothetical protein |

|
 0.33981 0.33981
| |
| Rv2023c | hypothetical protein |

|
 0.33944 0.33944
| |
| Rv1506c | hypothetical protein |


|
 0.3388 0.3388
| |
| Rv2355 | transposase |

|
 0.33823 0.33823
| |
| Rv3112 | molybdenum cofactor biosynthesis protein D moaD1 |

|
 0.3348 0.3348
| |
| Rv3378c | hypothetical protein |

|
 0.33365 0.33365
| |
| Rv0097 | oxidoreductase |

|
 0.33309 0.33309
| |
| Rv2027c | histidine kinase response regulator |

|
 0.3323 0.3323
| |
| Rv0943c | monooxygenase |

|
 0.33179 0.33179
| |
| Rv3110 | pterin-4-alpha-carbinolamine dehydratase moaB1 |

|
 0.33094 0.33094
| |
| Rv3108 | hypothetical protein |

|
 0.32286 0.32286
| |
| Rv2835c | sn-glycerol-3-phosphate transport membrane protein ABC transporter ugpA |

|
 0.32104 0.32104
| |
| Rv0026 | conserved hypothetical protein |

|
 0.32059 0.32059
| |
| Rv0893c | conserved hypothetical protein |

|
 0.31803 0.31803
| |
| Rv0564c | glycerol-3-phosphate dehydrogenase gpdA1 |

|
 0.31792 0.31792
| |
| Rv2336 | hypothetical protein |

|
 0.31791 0.31791
| |
| Rv2814c | transposase |

|
 0.31643 0.31643
| |
| Rv1504c | conserved hypothetical protein |

|
 0.31504 0.31504
| |
| Rv0457c | peptidase |

|
 0.31494 0.31494
| |
| Rv0899 | outer membrane protein A ompA |

|
 0.31254 0.31254
| |
| Rv0891c | transcriptional regulator |

|
 0.3114 0.3114
| |
| Rv2354 | transposase |

|
 0.31099 0.31099
| |
| Rv0160c | PE family protein |

|
 0.30772 0.30772
| |
| Rv2815c | transposase |

|
 0.30615 0.30615
| |
| Rv1763 | transposase |

|
 0.30492 0.30492
| |
| Rv0843 | dehydrogenase |

|
 0.30335 0.30335
| |
| Rv0488 | conserved membrane protein |

|
 0.3026 0.3026
| |
| Rv0393 | 13E12 repeat family protein |

|
 0.30254 0.30254
| |
| Rv2434c | conserved membrane protein |

|
 0.30176 0.30176
|