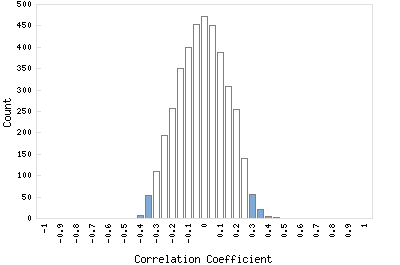
Correlation Catalog M. tuberculosis H37Rv: Rv0385 - monooxygenase
Distribution of gene correlation: Rv0385 vs. all genes
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Genes with negative correlation
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv3528c | hypothetical protein |

|
 -0.41164 -0.41164
| |
| Rv2954c | hypothetical protein |


|
 -0.39718 -0.39718
| |
| Rv3291c | transcriptional regulator, asnC-family |

|
 -0.38769 -0.38769
| |
| Rv2882c | ribosome recycling factor frr |

|
 -0.38354 -0.38354
| |
| Rv2301 | cutinase cut2 |

|
 -0.38174 -0.38174
| |
| Rv2953 | conserved hypothetical protein |

|
 -0.36867 -0.36867
| |
| Rv1078 | proline-rich antigen pra |

|
 -0.36169 -0.36169
| |
| Rv3224 | iron-regulated short-chain dehydrogenase/reductase |


|
 -0.35218 -0.35218
| |
| Rv0220 | esterase lipC |

|
 -0.35137 -0.35137
| |
| Rv0760c | conserved hypothetical protein |

|
 -0.34678 -0.34678
| |
| Rv3392c | cyclopropane-fatty-acyl-phospholipid synthase 1 cmaA1 |

|
 -0.34574 -0.34574
| |
| Rv3726 | dehydrogenase |


|
 -0.34269 -0.34269
| |
| Rv2140c | conserved hypothetical protein |

|
 -0.34109 -0.34109
| |
| Rv1824 | conserved membrane protein |

|
 -0.33979 -0.33979
| |
| Rv1884c | resuscitation-promoting factor rpfC |

|
 -0.3379 -0.3379
| |
| Rv3632 | conserved membrane protein |

|
 -0.33538 -0.33538
| |
| Rv3494c | MCE-family protein mce4F |

|
 -0.33449 -0.33449
| |
| Rv2348c | hypothetical protein |

|
 -0.33424 -0.33424
| |
| Rv2778c | conserved hypothetical protein |

|
 -0.33339 -0.33339
| |
| Rv3146 | NADH dehydrogenase I chain B nuoB |

|
 -0.33135 -0.33135
| |
| Rv3492c | MCE-associated protein |

|
 -0.33038 -0.33038
| |
| Rv3791 | short-chain type dehydrogenase/reductase |


|
 -0.32924 -0.32924
| |
| Rv3499c | MCE-family protein mce4A |

|
 -0.32853 -0.32853
| |
| Rv1547 | DNA polymerase III alpha chain dnaE1 |

|
 -0.32846 -0.32846
| |
| Rv1677 | lipoprotein dsbF |


|
 -0.32726 -0.32726
| |
| Rv1242 | conserved hypothetical protein |

|
 -0.32296 -0.32296
| |
| Rv3496c | MCE-family protein mce4D |

|
 -0.32281 -0.32281
| |
| Rv3498c | MCE-family protein mce4B |

|
 -0.32199 -0.32199
| |
| Rv2102 | conserved hypothetical protein |

|
 -0.32044 -0.32044
| |
| Rv3148 | NADH dehydrogenase I chain D nuoD |

|
 -0.31973 -0.31973
| |
| Rv0566c | conserved hypothetical protein |

|
 -0.31957 -0.31957
| |
| Rv3725 | oxidoreductase |


|
 -0.3193 -0.3193
| |
| Rv3631 | transferase |

|
 -0.31799 -0.31799
| |
| Rv3300c | conserved hypothetical protein |

|
 -0.31696 -0.31696
| |
| Rv3274c | acyl-CoA dehydrogenase fadE25 |

|
 -0.31566 -0.31566
| |
| Rv3576 | lipoprotein lppH |

|
 -0.31562 -0.31562
| |
| Rv1608c | peroxidoxin bcpB |

|
 -0.31541 -0.31541
| |
| Rv1614 | prolipoprotein diacylglyceryl transferases lgt |

|
 -0.31497 -0.31497
| |
| Rv2956 | conserved hypothetical protein |


|
 -0.31483 -0.31483
| |
| Rv3793 | membrane indolylacetylinositol arabinosyltransferase embC |

|
 -0.3148 -0.3148
| |
| Rv3152 | NADH dehydrogenase I chain H nuoH |

|
 -0.31465 -0.31465
| |
| Rv2775 | hypothetical protein |


|
 -0.31454 -0.31454
| |
| Rv0168 | hypothetical membrane protein yrbE1B |

|
 -0.31179 -0.31179
| |
| Rv0432 | periplasmic superoxide dismutase sodC |

|
 -0.31174 -0.31174
| |
| Rv3318 | succinate dehydrogenase flavoprotein subunit sdhA |

|
 -0.30983 -0.30983
| |
| Rv2220 | glutamine synthetase glnA1 |

|
 -0.30983 -0.30983
| |
| Rv0925c | conserved hypothetical protein |

|
 -0.30959 -0.30959
| |
| Rv3852 | histone-like protein hns |

|
 -0.30865 -0.30865
| |
| Rv2046 | lipoprotein lppI |

|
 -0.30799 -0.30799
| |
| Rv1703c | catechol-O-methyltransferase |


|
 -0.30708 -0.30708
| |
| Rv1612 | tryptophan synthase, beta subunit trpB |

|
 -0.30667 -0.30667
| |
| Rv3624c | hypoxanthine-guanine phosphoribosyltransferase hpt |

|
 -0.30641 -0.30641
| |
| Rv1984c | cutinase precursor cfp21 |

|
 -0.30602 -0.30602
| |
| Rv2560 | proline and glycine rich membrane protein |

|
 -0.30242 -0.30242
| |
| Rv1830 | conserved hypothetical protein |

|
 -0.30229 -0.30229
| |
| Rv0503c | cyclopropane-fatty-acyl-phospholipid synthase 2 cmaA2 |

|
 -0.30222 -0.30222
| |
| Rv2258c | transcriptional regulator |


|
 -0.30188 -0.30188
| |
| Rv2346c | Esat-6 like protein esxO |

|
 -0.30166 -0.30166
| |
| Rv3696c | glycerol kinase glpK |

|
 -0.30111 -0.30111
| |
| Rv2928 | thioesterase tesA |

|
 -0.30051 -0.30051
| |
| Rv3676 | transcriptional regulator, crp/fnr-family |

|
 -0.30028 -0.30028
| |
| Rv3633 | conserved hypothetical protein |

|
 -0.30008 -0.30008
|
Genes with positive correlation 
Categories Enriched  : None
: None
Gene  | Description | Links  | COG  | Correlation  |
|---|---|---|---|---|
| Rv0245 | oxidoreductase |

|
 0.51875 0.51875
| |
| Rv0389 | phosphoribosylglycinamide formyltransferase 2 purT |

|
 0.47787 0.47787
| |
| Rv0457c | peptidase |

|
 0.46385 0.46385
| |
| Rv0209 | hypothetical protein |

|
 0.44964 0.44964
| |
| Rv0627 | conserved hypothetical protein |

|
 0.4458 0.4458
| |
| Rv0793 | conserved hypothetical protein |

|
 0.44118 0.44118
| |
| Rv0121c | conserved hypothetical protein |

|
 0.4383 0.4383
| |
| Rv0473 | conserved membrane protein |

|
 0.41109 0.41109
| |
| Rv0775 | conserved hypothetical protein |

|
 0.39779 0.39779
| |
| Rv0037c | conserved membrane protein |




|
 0.39647 0.39647
| |
| Rv0417 | thiamin biosynthesis protein thiG |

|
 0.39533 0.39533
| |
| Rv0122 | hypothetical protein |

|
 0.39299 0.39299
| |
| Rv0557 | mannosyltransferase pimB |

|
 0.37697 0.37697
| |
| Rv0275c | transcriptional regulator, tetR-family |

|
 0.3725 0.3725
| |
| Rv1225c | conserved hypothetical protein |

|
 0.37246 0.37246
| |
| Rv0197 | oxidoreductase |

|
 0.36956 0.36956
| |
| Rv0915c | PPE family protein |

|
 0.36336 0.36336
| |
| Rv1395 | transcriptional regulator |

|
 0.36323 0.36323
| |
| Rv1929c | conserved hypothetical protein |

|
 0.36168 0.36168
| |
| Rv0826 | conserved hypothetical protein |

|
 0.36148 0.36148
| |
| Rv0089 | methyltransferase/methylase |


|
 0.36038 0.36038
| |
| Rv0123 | hypothetical protein |

|
 0.3579 0.3579
| |
| Rv0029 | conserved hypothetical protein |

|
 0.35705 0.35705
| |
| Rv0327c | cytochrome P450 135A1 cyp135A1 |

|
 0.35674 0.35674
| |
| Rv2310 | excisionase |

|
 0.35271 0.35271
| |
| Rv0330c | hypothetical protein |

|
 0.35265 0.35265
| |
| Rv0273c | transcriptional regulator |

|
 0.35217 0.35217
| |
| Rv1067c | PE-PGRS family protein |

|
 0.35072 0.35072
| |
| Rv0486 | mannosyltransferase |

|
 0.34864 0.34864
| |
| Rv0388c | PPE family protein |

|
 0.34595 0.34595
| |
| Rv3785 | hypothetical protein |

|
 0.34572 0.34572
| |
| Rv0875c | hypothetical exported protein |

|
 0.34509 0.34509
| |
| Rv0841c |

|
 0.34481 0.34481
| ||
| Rv2133c | conserved hypothetical protein |

|
 0.34219 0.34219
| |
| Rv1627c | nonspecific lipid-transfer protein |

|
 0.34197 0.34197
| |
| Rv2436 | ribokinase rbsK |

|
 0.34146 0.34146
| |
| Rv0152c | PE family protein |

|
 0.34037 0.34037
| |
| Rv2656c | phiRv2 phage protein |

|
 0.33951 0.33951
| |
| Rv1161 | respiratory nitrate reductase alpha chain narG |

|
 0.33939 0.33939
| |
| Rv1977 | conserved hypothetical protein |

|
 0.33522 0.33522
| |
| Rv2825c | conserved hypothetical protein |

|
 0.33494 0.33494
| |
| Rv1539 | lipoprotein signal peptidase lspA |

|
 0.33351 0.33351
| |
| Rv0537c | membrane protein |

|
 0.32759 0.32759
| |
| Rv0328 | transcriptional regulator, tetR/acrR-family |

|
 0.32584 0.32584
| |
| Rv3182 | conserved hypothetical protein |

|
 0.32532 0.32532
| |
| Rv1635c | conserved membrane protein |

|
 0.32355 0.32355
| |
| Rv0623 | conserved hypothetical protein |

|
 0.32272 0.32272
| |
| Rv0071 | maturase |

|
 0.32263 0.32263
| |
| Rv0617 | conserved hypothetical protein |

|
 0.32247 0.32247
| |
| Rv1939 | oxidoreductase |

|
 0.32186 0.32186
| |
| Rv0579 | conserved hypothetical protein |

|
 0.32056 0.32056
| |
| Rv0625c | conserved membrane protein |

|
 0.32039 0.32039
| |
| Rv1599 | histidinol dehydrogenase hisD |

|
 0.31975 0.31975
| |
| Rv0101 | peptide synthetase nrp |

|
 0.31914 0.31914
| |
| Rv0595c | conserved hypothetical protein |

|
 0.31879 0.31879
| |
| Rv0415 | thiamine biosynthesis oxidoreductase thiO |

|
 0.31729 0.31729
| |
| Rv0612 | conserved hypothetical protein |

|
 0.31719 0.31719
| |
| Rv2017 | transcriptional regulator |

|
 0.31597 0.31597
| |
| Rv3531c | hypothetical protein |

|
 0.31562 0.31562
| |
| Rv1628c | conserved hypothetical protein |

|
 0.31509 0.31509
| |
| Rv1894c | conserved hypothetical protein |

|
 0.3145 0.3145
| |
| Rv1585c | phiRv1 phage protein |

|
 0.3142 0.3142
| |
| Rv2497c | pyruvate dehydrogenase E1 component alpha subunit pdhA |

|
 0.31315 0.31315
| |
| Rv0378 | conserved glycine rich protein |

|
 0.31288 0.31288
| |
| Rv3180c | hypothetical alanine rich protein |

|
 0.31241 0.31241
| |
| Rv0477 | conserved secreted protein |

|
 0.31237 0.31237
| |
| Rv0340 | conserved hypothetical protein |

|
 0.31232 0.31232
| |
| Rv0326 | hypothetical protein |


|
 0.31064 0.31064
| |
| Rv0857 | conserved hypothetical protein |

|
 0.31041 0.31041
| |
| Rv1781c | 4-alpha-glucanotransferase malQ |

|
 0.30809 0.30809
| |
| Rv0369c | membrane oxidoreductase |

|
 0.30808 0.30808
| |
| Rv0754 | PE-PGRS family protein |

|
 0.30656 0.30656
| |
| Rv0077c | oxidoreductase |

|
 0.30511 0.30511
| |
| Rv3857c | membrane protein |

|
 0.30397 0.30397
| |
| Rv0471c | hypothetical protein |

|
 0.30232 0.30232
| |
| Rv2330c | lipoprotein lppP |

|
 0.30207 0.30207
| |
| Rv1351 | hypothetical protein |

|
 0.30163 0.30163
| |
| Rv3537 | dehydrogenase |

|
 0.30142 0.30142
| |
| Rv0597c | conserved hypothetical protein |

|
 0.30116 0.30116
| |
| Rv0271c | acyl-CoA dehydrogenase fadE6 |

|
 0.30081 0.30081
| |
| Rv0657c | conserved hypothetical protein |

|
 0.30035 0.30035
| |
| Rv2517c | hypothetical protein |

|
 0.30016 0.30016
| |
| Rv0215c | acyl-CoA dehydrogenase fadE3 |

|
 0.30003 0.30003
|